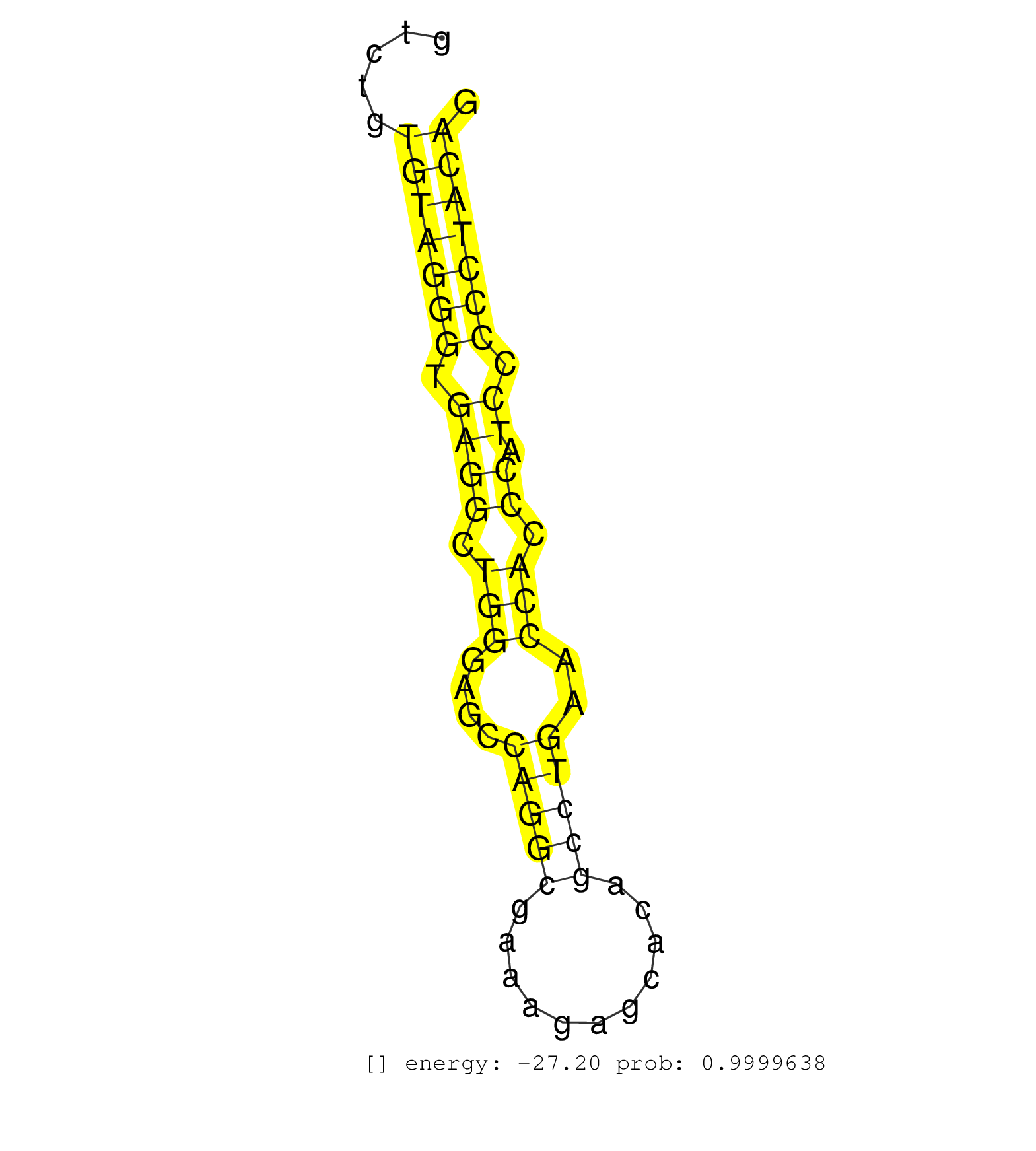
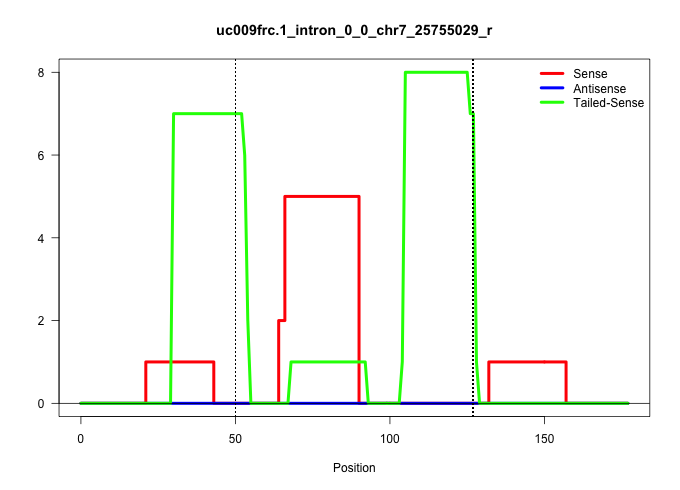
| Gene: Pra1 | ID: uc009frc.1_intron_0_0_chr7_25755029_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) OVARY |
(1) PIWI.ip |
(17) TESTES |
| CCCCTTCTTCTGGCTGGCCGGTGCTGGCTCTGCTGTCTTCTGGGTCCTGGGTGAGTACAGGGTCTGTGTAGGGTGAGGCTGGGAGCCAGGCGAAAGAGCACAGCCTGAACCACCCATCCCCCTACAGGAGCCACGCTGGTACTCATAGGCTCCCACGCTGCCTTCCACCAGATGGAG ..................................................................(((((((.((((.(((....(((((...........)))))..))).)).)).)))))))................................................... .............................................................62...............................................................127................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................TGAACCACCCATCCCCCTACAGt................................................. | 23 | t | 9.00 | 0.00 | - | 1.00 | - | 4.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 |
| ..................................................................TGTAGGGTGAGGCTGGGAGCCAGG....................................................................................... | 24 | 1 | 8.00 | 8.00 | 2.00 | - | - | - | 2.00 | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | 1.00 | - |
| ..............................TGCTGTCTTCTGGGTCCTGGGagc........................................................................................................................... | 24 | agc | 6.00 | 0.00 | 2.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................CTGTGTAGGGTGAGGCTGGGAGCCAGG....................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CTGGCTCTGCTGTCTTCTGGGTC................................................................................................................................... | 23 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TGTGTAGGGTGAGGCTGGGAGCCAGG....................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGAACCACCCATCCCCCTACAtt................................................. | 23 | tt | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..............................TGCTGTCTTCTGGGTCCTGGGagcc.......................................................................................................................... | 25 | agcc | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................GCTGGCTCTGCTGTCTTCTGGGTCCTGGGagc........................................................................................................................... | 32 | agc | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GGTGCTGGCTCTGCTG.............................................................................................................................................. | 16 | 6 | 1.17 | 1.17 | - | - | 1.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................CTGAACCACCCATCCCCCTACt................................................... | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGAACCACCCATCCCCCTACAc.................................................. | 22 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGGTACTCATAGGCTCCCACGCTGC................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TGCTGTCTTCTGGGTCCTGGGag............................................................................................................................ | 23 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................TGCTGGCTCTGCTGTCTTCTGG...................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................GTAGGGTGAGGCTGGGAGCCAGG....................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TGCTGTCTTCTGGGTCCTGGGaaa........................................................................................................................... | 24 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................TAGGGTGAGGCTGGGAGCCAGGCaa.................................................................................... | 25 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................TGAACCACCCATCCCCCTACAGta................................................ | 24 | ta | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................CCGGTGCTGGCTCTGCTGTCTTCTGG...................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACGCTGGTACTCATAGGCTCCCACG.................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGAACCACCCATCCCCCTACAGa................................................. | 23 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCCTTCTTCTGGCTGGCCGGTGCTGGCTCTGCTGTCTTCTGGGTCCTGGGTGAGTACAGGGTCTGTGTAGGGTGAGGCTGGGAGCCAGGCGAAAGAGCACAGCCTGAACCACCCATCCCCCTACAGGAGCCACGCTGGTACTCATAGGCTCCCACGCTGCCTTCCACCAGATGGAG ..................................................................(((((((.((((.(((....(((((...........)))))..))).)).)).)))))))................................................... .............................................................62...............................................................127................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|