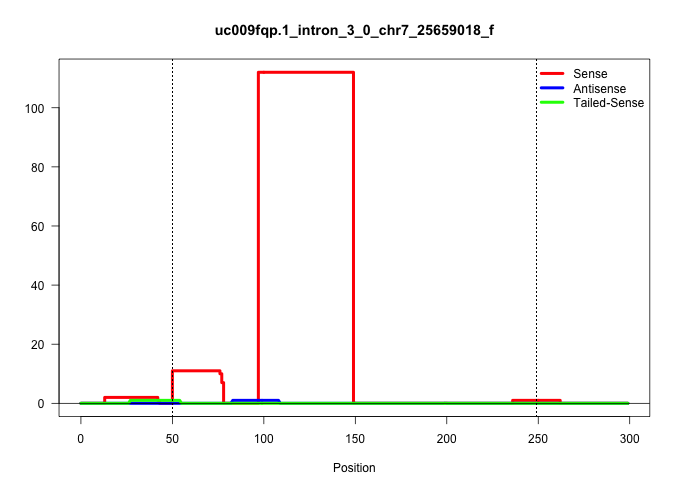
| Gene: Dmrtc2 | ID: uc009fqp.1_intron_3_0_chr7_25659018_f | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(1) PIWI.ip |
(13) TESTES |
| CTCAGAGTCCCCATGGAGCAGTCCCACTGGTACTGACACCCCCTGGGAAGGTAAGAGACCTGAGTCTGAGAGAGTGGCTCCTAGCCTGGCTCTAGCCCCAACTGTTTCCTCTGTGCTCCCTGGGTTCCAGCTGCAGCCCTGACTTGCAATGTGGGTGAACTAAAAAGGGACTTAAGAGCCGCCCACAGTTGCATGTCCCACCCTCATCCCAGAGCATGCTCACTTCAGCCTTGCCCTCTACCCTTGCAGGAGAACTATGGGCCACTGCTGCTCAGCCGCCCCCCGGAAGCCTTGCCTTT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................CCAACTGTTTCCTCTGTGCTCCCTGGGTTCCAGCTGCAGCCCTGACTTGCAA...................................................................................................................................................... | 52 | 1 | 112.00 | 112.00 | 112.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAGACCTGAGTCTGAGAGAGTGGC............................................................................................................................................................................................................................. | 28 | 1 | 7.00 | 7.00 | - | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 |
| ..................................................GTAAGAGACCTGAGTCTGAGAGAGTGG.............................................................................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - |
| .............TGGAGCAGTCCCACTGGTACTGACACCCCC................................................................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................TGGTACTGACACCCCCTGGGAAGGagaa.................................................................................................................................................................................................................................................... | 28 | agaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............TGGAGCAGTCCCACTGGTACTGACACCCC................................................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAGACCTGAGTCTGAGAGAGTG............................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................................................................TCTACCCTTGCAGGAGAACTATGGGC..................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| CTCAGAGTCCCCATGGAGCAGTCCCACTGGTACTGACACCCCCTGGGAAGGTAAGAGACCTGAGTCTGAGAGAGTGGCTCCTAGCCTGGCTCTAGCCCCAACTGTTTCCTCTGTGCTCCCTGGGTTCCAGCTGCAGCCCTGACTTGCAATGTGGGTGAACTAAAAAGGGACTTAAGAGCCGCCCACAGTTGCATGTCCCACCCTCATCCCAGAGCATGCTCACTTCAGCCTTGCCCTCTACCCTTGCAGGAGAACTATGGGCCACTGCTGCTCAGCCGCCCCCCGGAAGCCTTGCCTTT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................GCCTGGCTCTAGCCCCAACTGTTTCC.............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TCAGCCTTGCCCTCTctta............................................................ | 19 | ctta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |