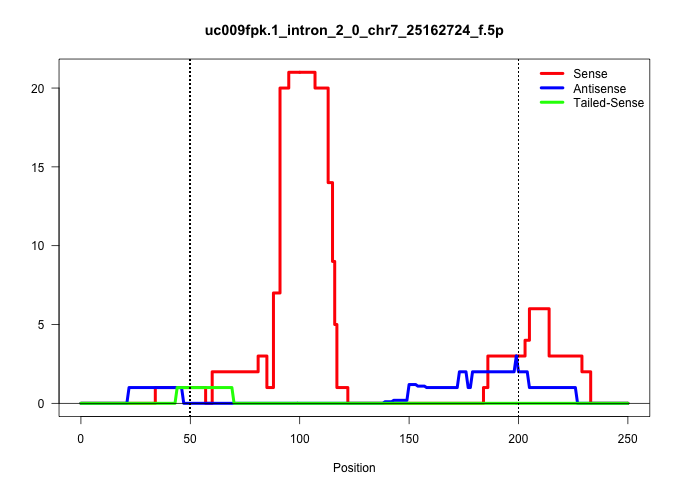
| Gene: Kcnn4 | ID: uc009fpk.1_intron_2_0_chr7_25162724_f.5p | SPECIES: mm9 |
 |
|
(6) OTHER.mut |
(1) PIWI.mut |
(13) TESTES |
| ACGCTGGGTCTCTGGCTCACCACAGCTTGGGTGCTGTCTGTGGCTGAGAGGTGAGGGGCCTGGAACACAGGGAGGAGACACCAGAGGCTACTGGATATGATGGGGCAGGCTGCAAGGCTGGGGGCCAGCCAACATCAGGGCAGGCTCACACTTTCCATCCTTCTTGGCTGAGAAATCTCAAGTCCTTGAGGTGATTCCCAACTTCTGAGCCACCCAGAACACACACACAGAGGGAAGGAGAGAGAGAGAG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................TACTGGATATGATGGGGCAGGCTGC......................................................................................................................................... | 25 | 1 | 6.00 | 6.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................TGGATATGATGGGGCAGGCTGCAA....................................................................................................................................... | 24 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGGATATGATGGGGCAGGCTGCAAG...................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................TGGATATGATGGGGCAGGCTGCAAGG..................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TGAGGTGATTCCCAACTTCTGAGCCACC.................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAGCCACCCAGAACACACACACAGAGG................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................TGGAACACAGGGAGGAGACACCAGA..................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................TATGATGGGGCAGGCTGCAAGGCTGGG................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................TGAGAGGTGAGGGGCCTGGAACACAa.................................................................................................................................................................................... | 26 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................TGTCTGTGGCTGAGAGGTGAGGG................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TCTGAGCCACCCAGAACACACACACA..................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................CAGAGGCTACTGGATATGATGGGGCA............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................................................CTTGAGGTGATTCCCAACTTCTGAGCCACC.................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ACGCTGGGTCTCTGGCTCACCACAGCTTGGGTGCTGTCTGTGGCTGAGAGGTGAGGGGCCTGGAACACAGGGAGGAGACACCAGAGGCTACTGGATATGATGGGGCAGGCTGCAAGGCTGGGGGCCAGCCAACATCAGGGCAGGCTCACACTTTCCATCCTTCTTGGCTGAGAAATCTCAAGTCCTTGAGGTGATTCCCAACTTCTGAGCCACCCAGAACACACACACAGAGGGAAGGAGAGAGAGAGAG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................AACTTCTGAGCCACCCAGAACACACACA....................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................AATCTCAAGTCCTTGAGGTGATTCCCA.................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................CTTTCCATCCTTCTTGGCTGAGAAATC......................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TCAGGGCAGGCTCACACgggc................................................................................................... | 21 | gggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................CAGCTTGGGTGCTGTCTGTGGCTGA........................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................AAGTCCTTGAGGTGATTCCCAACTTC............................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................GCAGGCTCACACTTT................................................................................................ | 15 | 10 | 0.10 | 0.10 | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - |
| ...............................................................................................................................................GCTCACACTTTCCAT............................................................................................ | 15 | 11 | 0.09 | 0.09 | - | 0.09 | - | - | - | - | - | - | - | - | - | - | - |