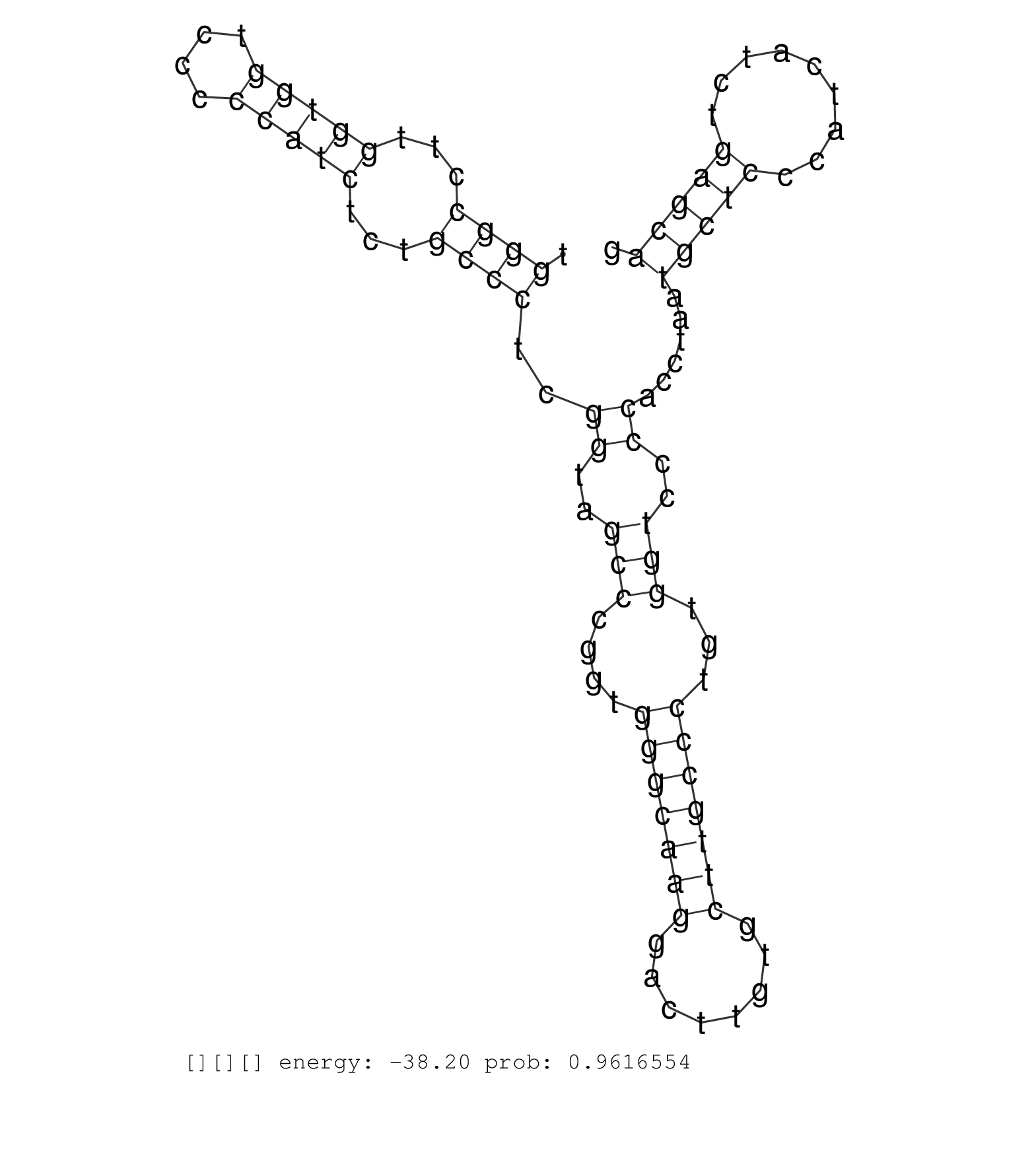
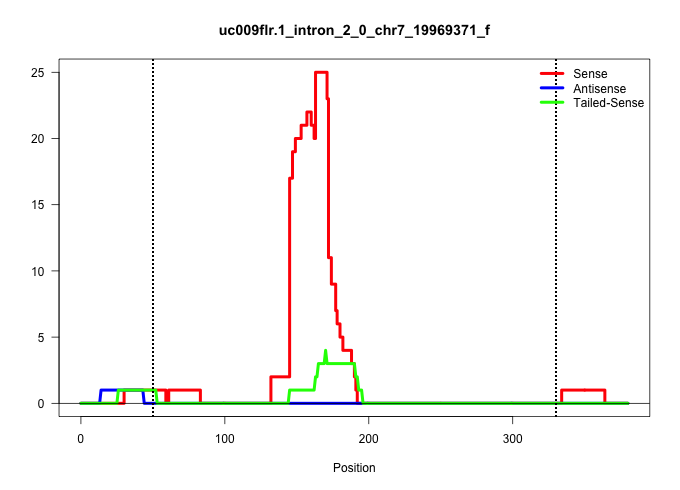
| Gene: Ercc2 | ID: uc009flr.1_intron_2_0_chr7_19969371_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(24) TESTES |
| GCACTGGGAAGACAGTGTCCCTATTGGCCCTGATTGTGGCCTATCAGCGGGTGAGTGGCCTTCTGAGTGCCTGGAGTGGGGGCTGAGGGGTGGGGCTGGATGGGGCTGAGGTCCCTGTCACCCACAGGCTTATCCGCTGGAGGTGACCAAACTTATCTACTGCTCACGGACCGTGCCAGAGATTGAGAAGGTGGGCAATGCAGATGGGACCCATGCTCTGCCCCCTCCCCTGGGCCTTGGTGGTCCCCCATCTCTGCCCTCGGTAGCCCGGTGGGCAAGGACTTGTGCTTGCCCTGTGGTCCCCACCTAATGCTCCCATCATCTGAGCAGGTCATAGAAGAGCTTCGTAAGCTACTCAGCTTCTACGAGCAGCAGGAGGG .......................................................................................................................................................................................................................................((((...(((((....)))))...))))..((..(((....(((((((........)))))))...)))..))......(((((.........)))))................................................... ......................................................................................................................................................................................................................................231................................................................................................330................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT3() Testes Data. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................ACCAAACTTATCTACTGCTCACGGACC................................................................................................................................................................................................................ | 27 | 1 | 12.00 | 12.00 | 4.00 | 3.00 | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................TCACGGACCGTGCCAGAGATTGAGA................................................................................................................................................................................................ | 25 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CAAACTTATCTACTGCTCACGGACCGT.............................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ACCAAACTTATCTACTGCTCACGGAC................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................GCCCTGATTGTGGCCTATCAGCGGGct....................................................................................................................................................................................................................................................................................................................................... | 27 | ct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TATCTACTGCTCACGGACCGTGCCA.......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TCACGGACCGTGCCAGAGATTGAGAAGGT............................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CCGTGCCAGAGATTGAGAAGGTcata........................................................................................................................................................................................ | 26 | cata | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TACTGCTCACGGACCGTGCCAGAGA...................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GCCCTGATTGTGGCCTATCAGCGGGctt...................................................................................................................................................................................................................................................................................................................................... | 28 | ctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................TCACGGACCGTGCCAGAGATTGAGAAGt............................................................................................................................................................................................. | 28 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................TCCGCTGGAGGTGACCAAACTTATCTACTG.......................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ACCAAACTTATCTACTGCTCACGGACCGTGCC........................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCCGCTGGAGGTGACCAAACTTATCTAC............................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TCTGAGTGCCTGGAGTGGGGGC......................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TCACGGACCGTGCCAGAGATTGAGAAGG............................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................TAGAAGAGCTTCGTAAGCTACTCAGCTTCT................ | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................ACGGACCGTGCCAGAGATTGAGAAGGTc........................................................................................................................................................................................... | 28 | c | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TCACGGACCGTGCCAGAGATTGAGAAG.............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TCACGGACCGTGCCAGA........................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................AACTTATCTACTGCTCACGGACCGTGCC........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TGATTGTGGCCTATCAGCGGGTGAGTGGC................................................................................................................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ACCAAACTTATCTACTGCTCACtgac................................................................................................................................................................................................................. | 26 | tgac | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGGATGGGGCTGAGGTCCCTGTC..................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................GGGGCTGAGGGGTGGGGC............................................................................................................................................................................................................................................................................................ | 18 | 6 | 0.17 | 0.17 | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCACTGGGAAGACAGTGTCCCTATTGGCCCTGATTGTGGCCTATCAGCGGGTGAGTGGCCTTCTGAGTGCCTGGAGTGGGGGCTGAGGGGTGGGGCTGGATGGGGCTGAGGTCCCTGTCACCCACAGGCTTATCCGCTGGAGGTGACCAAACTTATCTACTGCTCACGGACCGTGCCAGAGATTGAGAAGGTGGGCAATGCAGATGGGACCCATGCTCTGCCCCCTCCCCTGGGCCTTGGTGGTCCCCCATCTCTGCCCTCGGTAGCCCGGTGGGCAAGGACTTGTGCTTGCCCTGTGGTCCCCACCTAATGCTCCCATCATCTGAGCAGGTCATAGAAGAGCTTCGTAAGCTACTCAGCTTCTACGAGCAGCAGGAGGG .......................................................................................................................................................................................................................................((((...(((((....)))))...))))..((..(((....(((((((........)))))))...)))..))......(((((.........)))))................................................... ......................................................................................................................................................................................................................................231................................................................................................330................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT3() Testes Data. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................................................................................................GCAGGTCATAGAAGcgag........................................ | 18 | cgag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................................TGGGCCTTGGTGGTtgg........................................................................................................................................ | 17 | tgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............GTGTCCCTATTGGCCCTGATTGTGGCCTAT................................................................................................................................................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................AGGCTTATCCGctga.................................................................................................................................................................................................................................................... | 15 | ctga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |