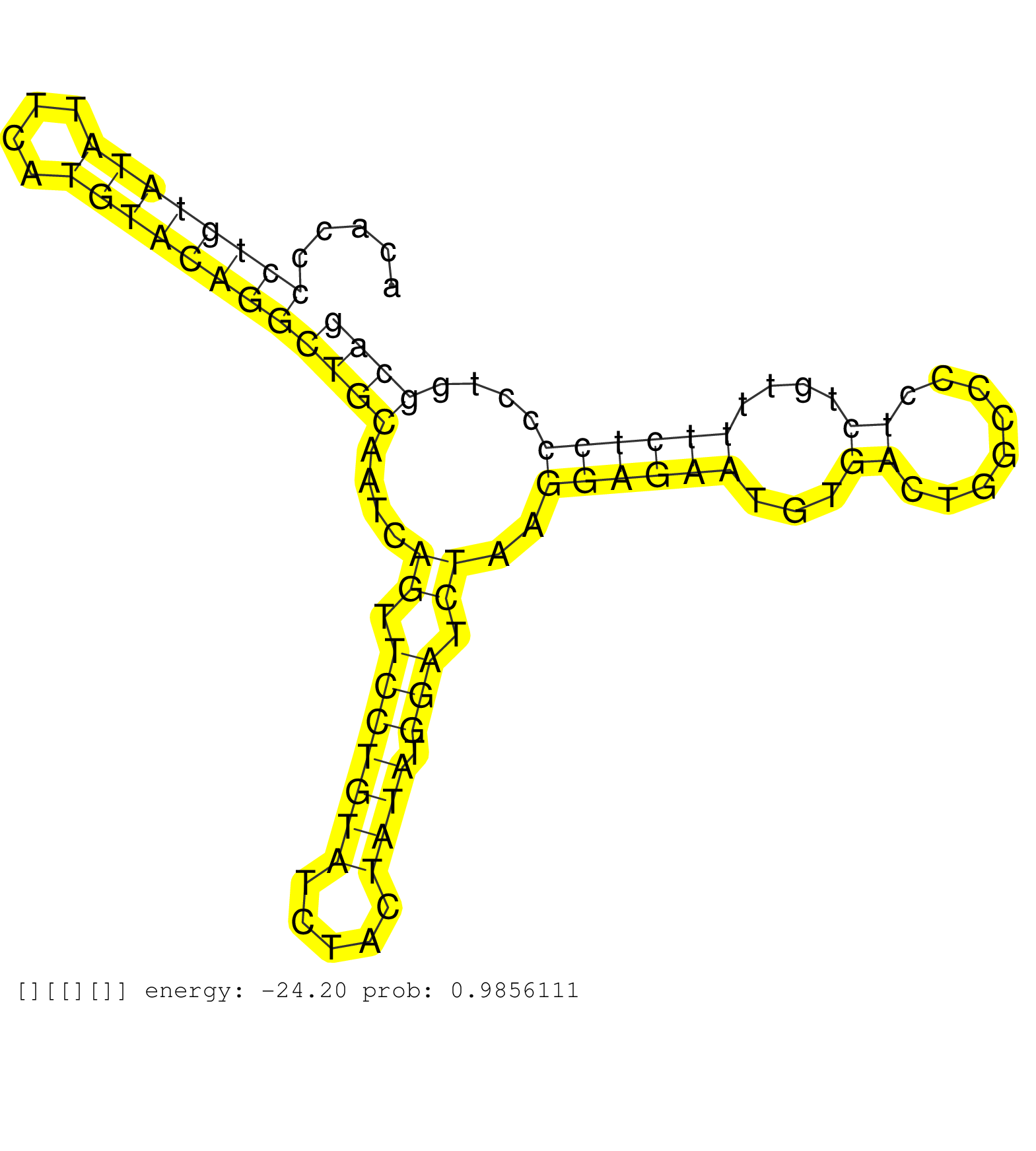
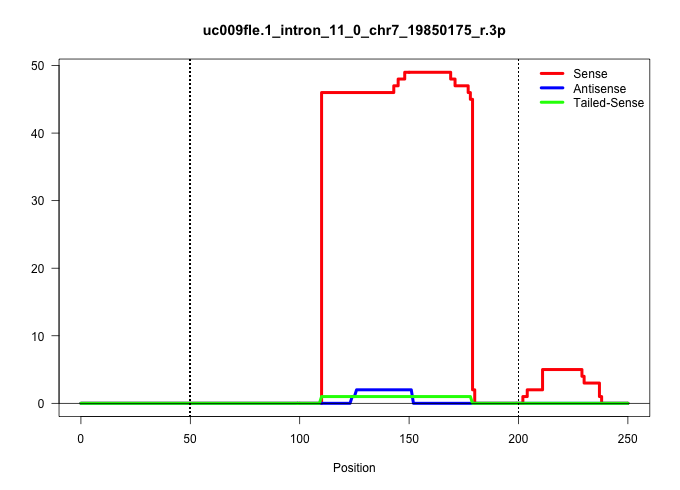
| Gene: Vasp | ID: uc009fle.1_intron_11_0_chr7_19850175_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| CAAACACGCTACTATTGAGCCACACCCCCAGCCCCTCATTGGGGTTTCTAGTCAGGGTCTCTAACCACTGAGCAACACTCCCAGTCATGCATGTACAGCCACACCCCTGTATATTCATGTACAGGCTGCAATCAGTTCCTGTATCTACTATATGGATCTAAGGAGAATGTGACTGGCCCCTCTGTTTTCTCCCCTGGCAGCGAGACGGTCATCTGTTCCAGCCGGGCTACTGTGATGCTTTATGATGACA .........................................................................................................((((((((....))))))))((((....((.(((((((.....)))).))).))..((((((...((........))....))))))....)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................ATATTCATGTACAGGCTGCAATCAGTTCCTGTATCTACTATATGGATCTAAG........................................................................................ | 52 | 1 | 48.00 | 48.00 | 48.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCTGTTCCAGCCGGGCTACTGTGATG............. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................TATATGGATCTAAGGAGAATGTGACTGGC......................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............TGAGCCACACCCCCAGCCCCTCAaaag................................................................................................................................................................................................................ | 27 | aaag | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................AGGAGAATGTGACTGGCCCCTCTG.................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................................AGACGGTCATCTGTTCCAGCCGGGCTAC.................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CCGGGCTACTGTGATG............. | 16 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................TCTACTATATGGATCTAAGGAGAATG................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................TACTATATGGATCTAAGGAGAATGTG............................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCTGTTCCAGCCGGGCTACTGTGATGC............ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................ACGGTCATCTGTTCCAGCCGGGCTA..................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| CAAACACGCTACTATTGAGCCACACCCCCAGCCCCTCATTGGGGTTTCTAGTCAGGGTCTCTAACCACTGAGCAACACTCCCAGTCATGCATGTACAGCCACACCCCTGTATATTCATGTACAGGCTGCAATCAGTTCCTGTATCTACTATATGGATCTAAGGAGAATGTGACTGGCCCCTCTGTTTTCTCCCCTGGCAGCGAGACGGTCATCTGTTCCAGCCGGGCTACTGTGATGCTTTATGATGACA .........................................................................................................((((((((....))))))))((((....((.(((((((.....)))).))).))..((((((...((........))....))))))....)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................CTGTTCCAGCCGGGCTACTGTGATGCTTt.......... | 29 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................GCTGCAATCAGTTCCTGTATCTACTATA.................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GCTGCAATCAGTTCCTGTATCTACTA.................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................TGCAATCAGTTCCTGTATCTACTATA.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................TCTCTAACCACTGAaggc................................................................................................................................................................................... | 18 | aggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |