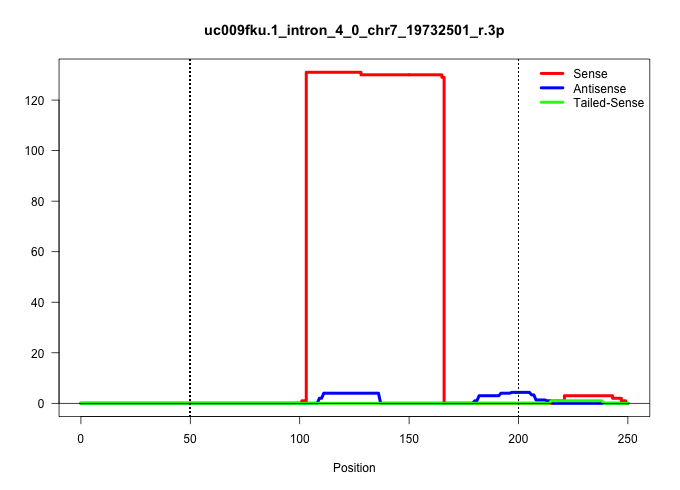
| Gene: Qpctl | ID: uc009fku.1_intron_4_0_chr7_19732501_r.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| CATGGCAAGTTTTGTTGCTACCGAGAAGTATGGTGGTTCTACATCTGGCTCCCAGCAGAAGTCAGTGTCGAGGGAACAACATAAACCAGAGTGGGACTTTCTAGTCACCCCGACTGCTCTGCGTGGCTCCAGTCACAAGGCCCTTCTCCCCACCTCCTCCCCAGTCCCTGTCCACCCTCCTACCTCTCTCTTGCCACTAGTTCCTGGAGGCTACGTTGCAGTCCCTGTCGGCAGGCTGGCATGTTGAACT .....................................................................................................(((((.....)))))..(((.(((((....(((....)))....................(((...)))......................)))))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................GTCACCCCGACTGCTCTGCGTGGCTCCAGTCACAAGGCCCTTCTCCCCACCT............................................................................................... | 52 | 1 | 132.00 | 132.00 | 132.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................ACGTTGCAGTCCCTGTCGGCAG................ | 22 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................CCCCAGTCCCTGTCCACC.......................................................................... | 18 | 3 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TCCCTGTCGGCAGGCTGGCATGTTGA... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TAGTCACCCCGACTGCTCTGCGTGGCT.......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TTGCAGTCCCTGTCGGCAGGCTGa........... | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGCAGTCCCTGTCGGCAGGCTGGCATG....... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TCCCTGTCGGCAGGCTGGCATGTTGAAC. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CATGGCAAGTTTTGTTGCTACCGAGAAGTATGGTGGTTCTACATCTGGCTCCCAGCAGAAGTCAGTGTCGAGGGAACAACATAAACCAGAGTGGGACTTTCTAGTCACCCCGACTGCTCTGCGTGGCTCCAGTCACAAGGCCCTTCTCCCCACCTCCTCCCCAGTCCCTGTCCACCCTCCTACCTCTCTCTTGCCACTAGTTCCTGGAGGCTACGTTGCAGTCCCTGTCGGCAGGCTGGCATGTTGAACT .....................................................................................................(((((.....)))))..(((.(((((....(((....)))....................(((...)))......................)))))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................GACTGCTCTGCGTGGCTCCAGTCACA................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................CCGACTGCTCTGCGTGGCTCCAGTCACA................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CCTCTCTCTTGCCACTAGTTCCTGGA.......................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................GCCACTAGTTCCTGGAGGCTACGT.................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................ACCCTCCTACCTCTtcat............................................................... | 18 | tcat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................................TCCACCCTCCTACCTCTt............................................................... | 18 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................CTACCGAGAAGTATGGTGGTTCTACATCTGa........................................................................................................................................................................................................... | 31 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................TACCTCTCTCTTGCCACTAGTTCCTG............................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGTTGCTACCGAGAAGTATGGTGGTTCTAt................................................................................................................................................................................................................. | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................................TAGTTCCTGGAGGCTA..................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................CCCCAGTCCCTGTCCA............................................................................ | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |