
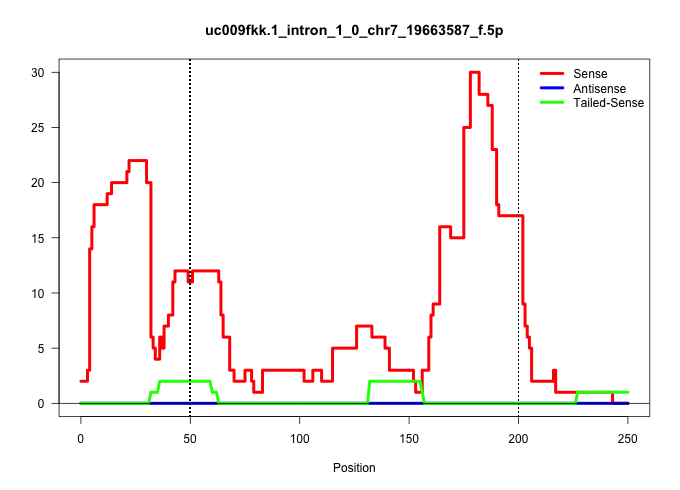
| Gene: Dmwd | ID: uc009fkk.1_intron_1_0_chr7_19663587_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| AGTATCTGGACCTCATCAAGAAAGACACCAGCAAGCTATTCAATGAAGAGGTGACTAAGCACGCGAGCCCTCCCCTTCTCACCTACCTACCCACCCCTGCCTGCACTAAGCTGCCTGGAAACAGAATAGGCTTGAAAGGGGCAGGAGGAGGGAGAGGGGACAAGTGACAGACAGCTACTGATGTGCCATTGTGCTTACTATTTCCCAGTTTTCTGCTGTGCCAGCCCTATGTTGGAAGAATGCTTTACTG ..............................................................................................................(((((((....)))......(((....)))))))..(((((.((..((.((((...(((..(((...))).)))....)))).))..)).)))))............................................. ..............................................................................................................111............................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....TCTGGACCTCATCAAGAAAGACACCAGC.......................................................................................................................................................................................................................... | 28 | 1 | 12.00 | 12.00 | - | 2.00 | - | - | 5.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | - |
| ...............................................................................................................................................................................TACTGATGTGCCATTGTGCTTACTATT................................................ | 27 | 1 | 8.00 | 8.00 | 4.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................TGACAGACAGCTACTGATGTGCCATT............................................................ | 26 | 1 | 5.00 | 5.00 | - | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................ATGAAGAGGTGACTAAGCACGCGAGC...................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGATGTGCCATTGTGCTTACTATTTCCC............................................ | 28 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGGAAACAGAATAGGCTTGAAAGGGG............................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CAAGTGACAGACAGCTACTGATGTGCCA.............................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................GGGACAAGTGACAGACAGCTACTGAT.................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................ACAAGTGACAGACAGCTACTGATGTGCCA.............................................................. | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGTGCTTACTATTTCCCAGTTTTCTGCT................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TATTCAATGAAGAGGTGACTAAGCACGC.......................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................TACTGATGTGCCATTGTGCTTACTATTT............................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CTGGACCTCATCAAGAAAGACACCAGC.......................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGAAAGGGGCAGGAGGAGGGAGAGt............................................................................................. | 25 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TCAATGAAGAGGTGACTAAGCACG........................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGATGTGCCATTGTGCTTACTATTTC.............................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TATGTTGGAAGAATGCTTTACTGtta | 26 | tta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TACCTACCCACCCCTGCCTGCACTAAG............................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................ACAAGTGACAGACAGCTACTGATGTGC................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGGAGGAGGGAGAGGGGACAAGTGACA................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......TGGACCTCATCAAGAAAGACACCAGCAA........................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................AAGACACCAGCAAGCTATTCAATGAAGA......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGACAGACAGCTACTGATGTGCCAT............................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................AAGTGACAGACAGCTACTGATGTGCCAT............................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TAGGCTTGAAAGGGGCAGGAGGAGGG.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGCCTGGAAACAGAATAGGCTTGAAAGG............................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............ATCAAGAAAGACACCAGCAAGCT..................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGACAGACAGCTACTGATGTGCCATTG........................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TTCAATGAAGAGGTGACTAAGCACGCG......................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....TCTGGACCTCATCAAGAAAGACACCAGCA......................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................TGAAGAGGTGACTAAGCACGCGAGCCC.................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGTGCCAGCCCTATGTTGGAAGAATGC....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...ATCTGGACCTCATCAAGAAAGACACCAGC.......................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................TGACTAAGCACGCGAGCCCTCCCCTTC............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TACCTACCCACCCCTGCCTGCACTAAGC........................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TTCAATGAAGAGGTGACTAAGCACGC.......................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGACTAAGCACGCGAGCCCTCCCCTTCT........................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGAAAGGGGCAGGAGGAGGGAGAa.............................................................................................. | 24 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGGAAACAGAATAGGCTTGAAAGGGGC............................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TAAGCTGCCTGGAAACAGAATAGGCTT..................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......TGGACCTCATCAAGAAAGACACCAGC.......................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................AAGCTATTCAATGAAGAGGTGACTAggc.............................................................................................................................................................................................. | 28 | ggc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TAGGCTTGAAAGGGGCAGGAGGAGGGA................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................TGATGTGCCATTGTGCTTACTATTTCC............................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TTCTCACCTACCTACCCACCCCTGCCT.................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TATTCAATGAAGAGGTGACTAAGacgc........................................................................................................................................................................................... | 27 | acgc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AGACACCAGCAAGCTATTCAATGAAGAGG....................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................CAATGAAGAGGTGACTAAGCACGCG......................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............TCATCAAGAAAGACACCAGCAAGCTAT................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| AGTATCTGGACCTCATCAAGAAAGACACCAGCAAGCTATTCAATGAAGAGGTGACTAAGCACGCGAGCCCTCCCCTTCTCACCTACCTACCCACCCCTGCCTGCACTAAGCTGCCTGGAAACAGAATAGGCTTGAAAGGGGCAGGAGGAGGGAGAGGGGACAAGTGACAGACAGCTACTGATGTGCCATTGTGCTTACTATTTCCCAGTTTTCTGCTGTGCCAGCCCTATGTTGGAAGAATGCTTTACTG ..............................................................................................................(((((((....)))......(((....)))))))..(((((.((..((.((((...(((..(((...))).)))....)))).))..)).)))))............................................. ..............................................................................................................111............................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|