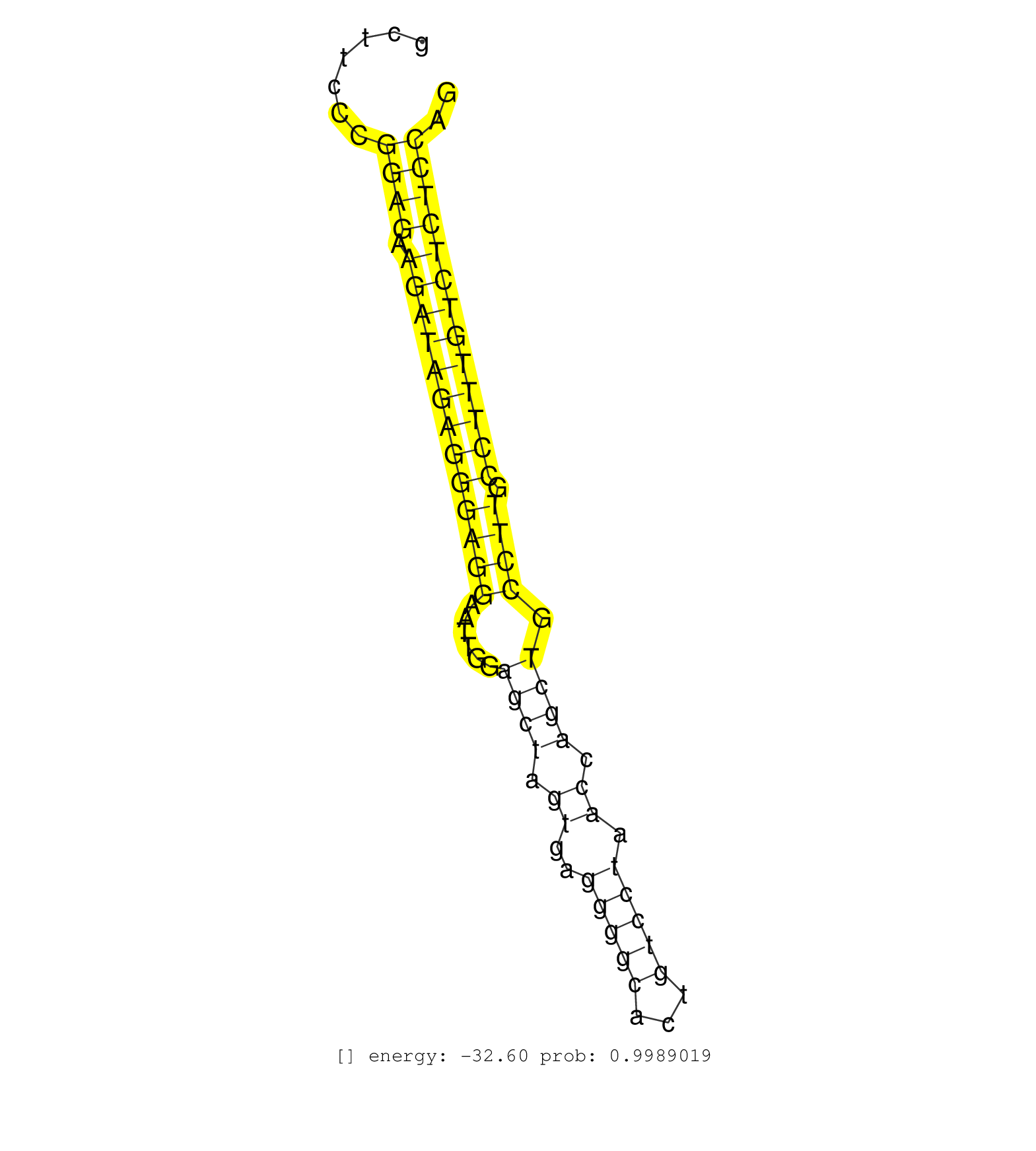

| Gene: Strn4 | ID: uc009fie.1_intron_16_0_chr7_17425322_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(8) TESTES |
| TCGCCCTGGCTGCACAGTGTCAGGGAGCGGGGCCAGGCAGTGGGGCTGAGGTAAGGATCAGGGCCATTTGTGTTCTAGATAGGAATGCAGACACCCAGTGTCCTTGCCCCAGAAGACACCCCTATACCCACCCCACCTCCATGATCCACTTGCCTGTCAGTTGGGTTGTGCCAAAGCCCACAGTCCTGTGGGATAAATGAGGCTTCCCGGAGAAGATAGAGGGAGGAATTGGAGCTAGTGAGGGGCACTGTCCTAACCAGCTGCCTTGCCTTTGTCTCTCCAGGCAGCCCCCAGCCTCACCTTCAGAGGCTCTGAGTCTTGGCCTCTTTCATC ................................................................................................................................................................................................................((((.(((((((((((((......((((.((..(((((...))))).)).)))).)))).))))))))))))).................................................... .........................................................................................................................................................................................................202..............................................................................283................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........TGCACAGTGTCAGGGAGCGGGGCCAG......................................................................................................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 |
| .........................................GGGGCTGAGGTAAGcgag.................................................................................................................................................................................................................................................................................. | 18 | cgag | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - |
| ................................CCAGGCAGTGGGGCTGAG........................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................................CCGGAGAAGATAGAGGGAGGAATTGG..................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CCGGAGAAGATAGAGGGAGGAATTGGA.................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..........TGCACAGTGTCAGGGAGCGGGGCCAGG........................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................TGCCTTGCCTTTGTCTCTCCAGt................................................. | 23 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................................................................................................................ACCTTCAGAGGCTCTGAGTCTTGG........... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| TCGCCCTGGCTGCACAGTGTCAGGGAGCGGGGCCAGGCAGTGGGGCTGAGGTAAGGATCAGGGCCATTTGTGTTCTAGATAGGAATGCAGACACCCAGTGTCCTTGCCCCAGAAGACACCCCTATACCCACCCCACCTCCATGATCCACTTGCCTGTCAGTTGGGTTGTGCCAAAGCCCACAGTCCTGTGGGATAAATGAGGCTTCCCGGAGAAGATAGAGGGAGGAATTGGAGCTAGTGAGGGGCACTGTCCTAACCAGCTGCCTTGCCTTTGTCTCTCCAGGCAGCCCCCAGCCTCACCTTCAGAGGCTCTGAGTCTTGGCCTCTTTCATC ................................................................................................................................................................................................................((((.(((((((((((((......((((.((..(((((...))))).)).)))).)))).))))))))))))).................................................... .........................................................................................................................................................................................................202..............................................................................283................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|