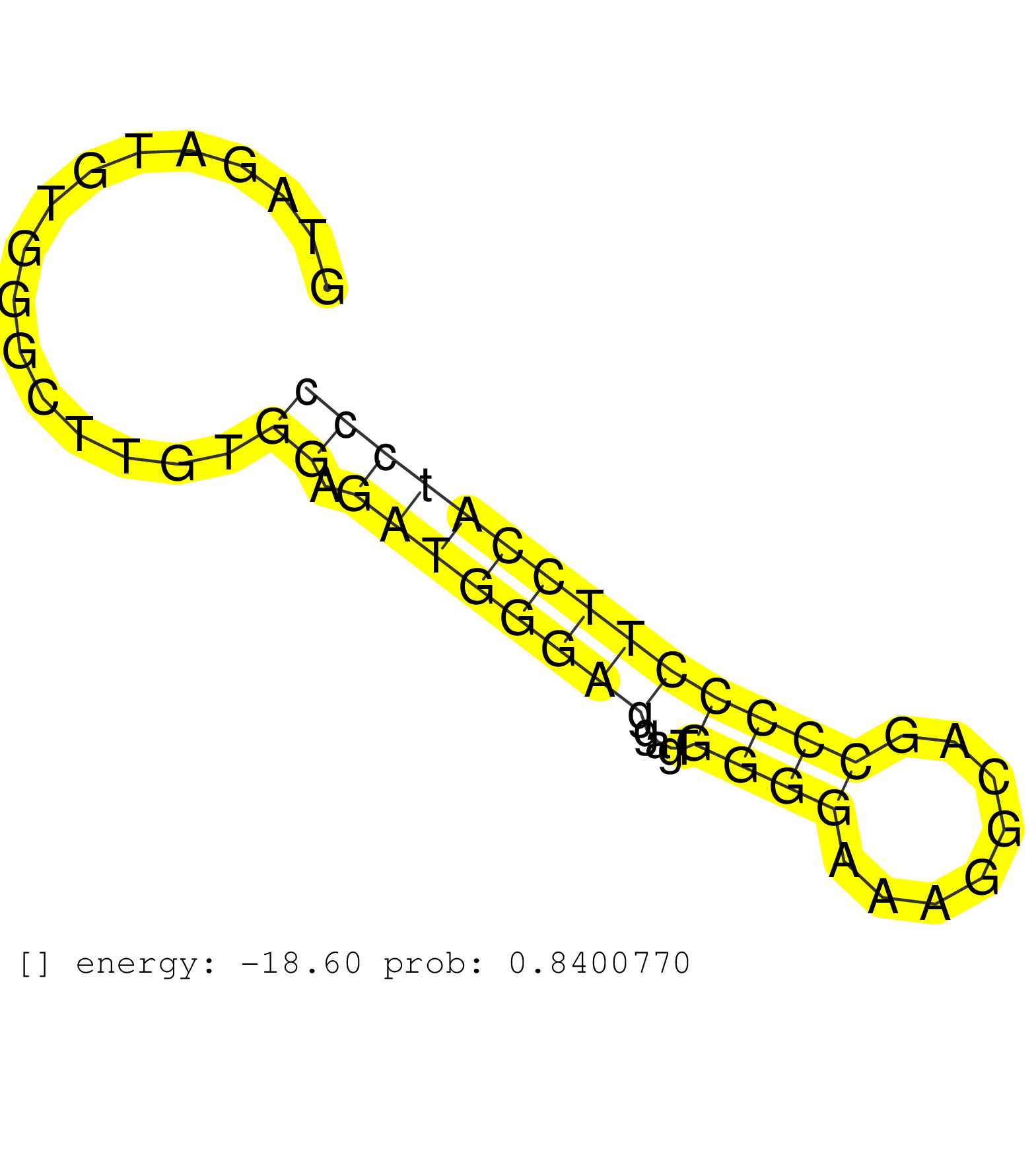

| Gene: Saps1 | ID: uc009eyb.1_intron_9_0_chr7_4591214_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(25) TESTES |
| ATGACCGCCTCAAGGAGTTCAACTTCCCTGAGGAGGCTGTATTGCAACAGGTAGATGTGGGCTTGTGGAGATGGGAGGAGTGGGGAAAGGCAGCCCCCTTCCATCCCTGACCCCCGTGCCTCTTGTTAGGCCTTCATGGACTTCCAGATGCAACGCATGACCTCAGCCTTCATTGACCA ..................................................................((.((((((((....((((........))))))))))))))........................................................................ ..................................................51......................................................107...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGATGTGGGCTTGTGGAGATGGGA....................................................................................................... | 26 | 1 | 73.00 | 73.00 | 16.00 | 8.00 | 10.00 | 4.00 | 5.00 | 7.00 | 4.00 | 3.00 | 1.00 | 3.00 | 4.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 |
| ..................................................GTAGATGTGGGCTTGTGGAGATGGG........................................................................................................ | 25 | 1 | 57.00 | 57.00 | 10.00 | 4.00 | 8.00 | 6.00 | 7.00 | 8.00 | 2.00 | 3.00 | - | 1.00 | 1.00 | - | 3.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAGATGG......................................................................................................... | 24 | 1 | 18.00 | 18.00 | 2.00 | 2.00 | 2.00 | 6.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TCATGGACTTCCAGATGCAACGCATGAC.................. | 28 | 1 | 8.00 | 8.00 | - | 4.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................TCATGGACTTCCAGATGCAACGCATGACC................. | 29 | 1 | 6.00 | 6.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAGATGGGAt...................................................................................................... | 27 | t | 5.00 | 73.00 | - | - | 1.00 | - | - | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGATGTGGGCTTGTGGAGATGGGA....................................................................................................... | 25 | 1 | 5.00 | 5.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................TGGACTTCCAGATGCAACGCATGACCT................ | 27 | 1 | 5.00 | 5.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAGATGGGAG...................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGGACTTCCAGATGCAACGCATGACCTC............... | 28 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAGATGGGt....................................................................................................... | 26 | t | 3.00 | 57.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TCATGGACTTCCAGATGCAACGCATGA................... | 27 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAGATG.......................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................CCTGAGGAGGCTGTATTGCAACAGGc............................................................................................................................... | 26 | c | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAGATGGGAa...................................................................................................... | 27 | a | 2.00 | 73.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TCATGGACTTCCAGATGCAACGCATGACCT................ | 30 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGGGGAAAGGCAGCCCCCTTCCA............................................................................ | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................GATGCAACGCATGACCTCAGCCTTCATT..... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTCATGGACTTCCAGATGCAACGCATGA................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGGCTGTATTGCAACAG................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TGGAGATGGGAGGAGTGGGGta............................................................................................ | 22 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAG............................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGCAACGCATGACCTCAGCCTT......... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................ACTTCCCTGAGGAGGCTGTATTGCAA.................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................ATGGACTTCCAGATGCAACGCATGgcc................. | 27 | gcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................TAGATGTGGGCTTGTGGAGATGGGAG...................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTCATGGACTTCCAGATGCAACGCATGACCT................ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CATGGACTTCCAGATGCAACGCATGACC................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................ATGGACTTCCAGATGCAACGCATGACCTC............... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGGACTTCCAGATGCAACGCATGACC................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGATGTGGGCTTGTGGAGATGGG........................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAGATGGGc....................................................................................................... | 26 | c | 1.00 | 57.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................TCCCTGAGGAGGCTGTATTGCAACAG................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTCATGGACTTCCAGATGCAACGC....................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGATGTGGGCTTGTGGAGATGGGAa...................................................................................................... | 26 | a | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAGATGaga....................................................................................................... | 26 | aga | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TCCAGATGCAACGCATGACCTCA.............. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CATGGACTTCCAGATGCAACGCATGACCTC............... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................GATGTGGGCTTGTGGAGATGGGAG...................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGATGTGGGCTTGTGGAGATGGGAGtt.................................................................................................... | 27 | tt | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTCATGGACTTCCAGATGCAACGCATG.................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGATGTGGGCTTGTGGAGATGtg........................................................................................................ | 25 | tg | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGATGTGGGCTTGTGGAGATGGGAG...................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATGACCGCCTCAAGGAGTTCAACTTCCCTGAGGAGGCTGTATTGCAACAGGTAGATGTGGGCTTGTGGAGATGGGAGGAGTGGGGAAAGGCAGCCCCCTTCCATCCCTGACCCCCGTGCCTCTTGTTAGGCCTTCATGGACTTCCAGATGCAACGCATGACCTCAGCCTTCATTGACCA ..................................................................((.((((((((....((((........))))))))))))))........................................................................ ..................................................51......................................................107...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................CAGCCTTCATTGAgtga... | 17 | gtga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |