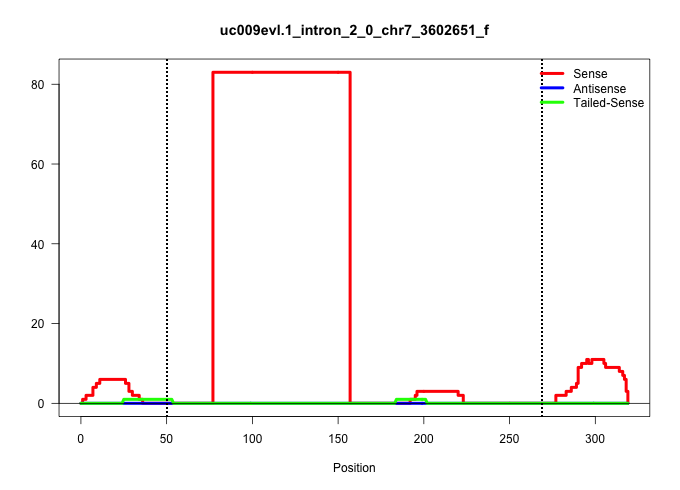
| Gene: Cnot3 | ID: uc009evl.1_intron_2_0_chr7_3602651_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
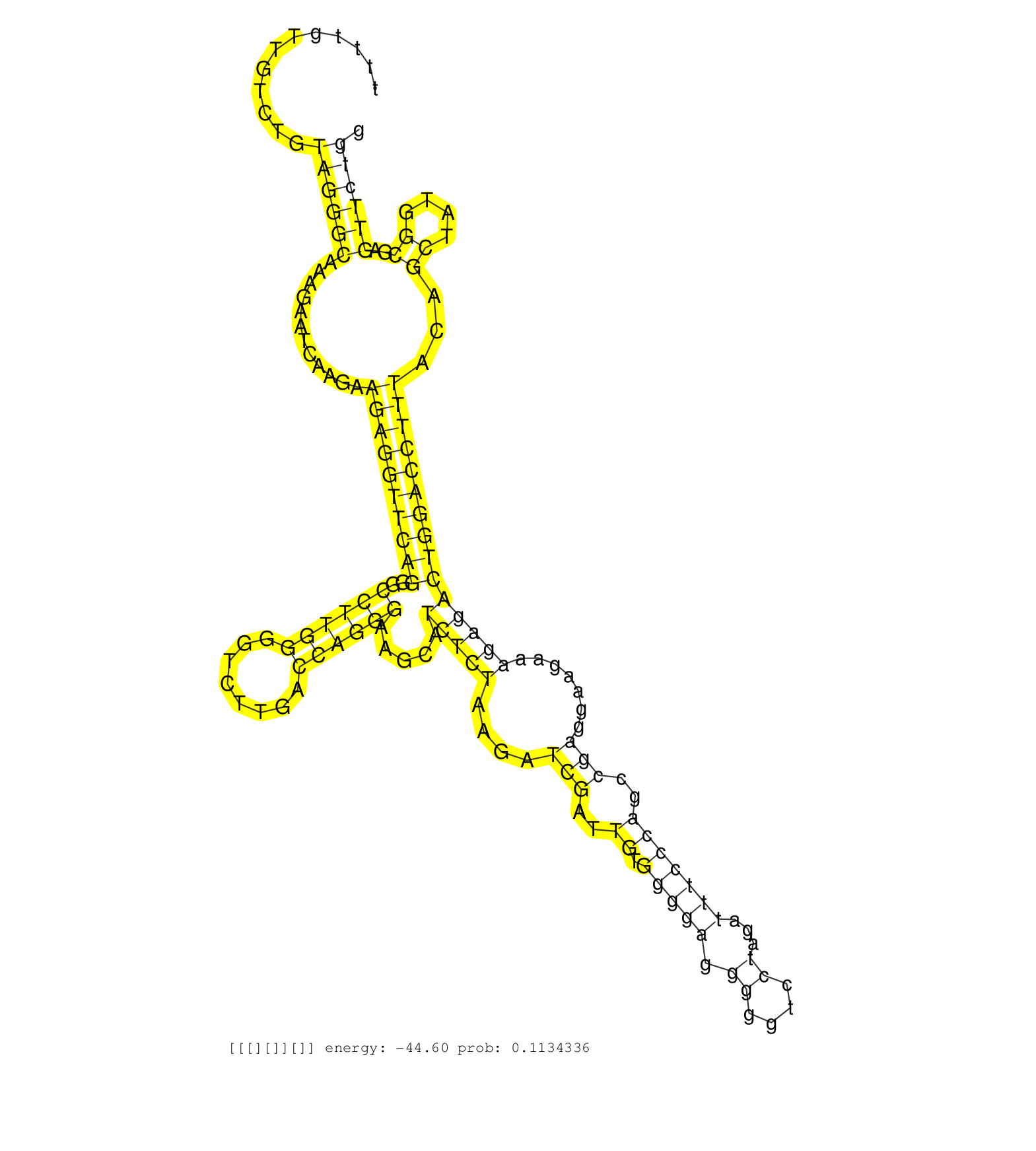
| TCAAGAAGGTGTCCGAAGGGGTGGAGCAGTTTGAAGATATTTGGCAAAAGGTAAGGACCTAGGAACCTTAAATTTTGTTGTCTGTAGGGCAAAGAATCAAGAAGAGGTTCAGGGCCTTGGGGTCTTGACCAGGGAAGCATCTCTAAGATCGATTGTGGGGAGGGGGTCCTAGATTTCCCAGCCGAGGAAGAAAGAGACTGGACCTTTACAGCTATGGCGAGTTCTGGGCAGCCACTTTCACATTGACTGCTGAATTCTTCATCCCTCAGCTCCACAATGCAGCCAACGCGAACCAGAAAGAAAAGTATGAGGCTGACCT ....................................................................................((((((............((((((((((..((((((........)))))).....(((((....(((..((.(((((.((....))...)))))))..))).......)))))))))))))))...((....))..))))))............................................................................................. ........................................................................73........................................................................................................................................................227.......................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................TTGTCTGTAGGGCAAAGAATCAAGAAGAGGTTCAGGGCCTTGGGGTCTTGAC.............................................................................................................................................................................................. | 52 | 1 | 84.00 | 84.00 | 84.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................GGAAGAAAGAGACTGGACC................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .......GGTGTCCGAAGGGGTGGAGCA................................................................................................................................................................................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................AACCAGAAAGAAAAGTATGAGGCTGACCT | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................TGCAGCCAACGCGAACCAG....................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................GAACCAGAAAGAAAAGTATGAGGCTGACC. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................CCAGAAAGAAAAGTATGAGGCTGACC. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................ACTGGACCTTTACAGCTATGGCGAGTT................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...AGAAGGTGTCCGAAGGGGTGGAGCAGT................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................AGAGACTGGACCTTTACAGCTATGGCGA................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................GACTGGACCTTTACAGCTATGGCGAGTT................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................TGCAGCCAACGCGAACCAGAAAGAAAAG.............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........TCCGAAGGGGTGGAGCAGTTTGAAG........................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .CAAGAAGGTGTCCGAAGGGGTGGAG..................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................GTCTGTAGGGCAAAGAATCAAGA......................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................CGCGAACCAGAAAGAAAAGTATGAGGCT..... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................AGGAAGAAAGAGACTatt..................................................................................................................... | 18 | att | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................AGAAAAGTATGAGGCTGACCT | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................AACCAGAAAGAAAAGTATGAGGCTGACC. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................GCAGTTTGAAGATATTTGGCAAAAGctcc......................................................................................................................................................................................................................................................................... | 29 | ctcc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................AACCAGAAAGAAAAGTATGAGGCTGAC.. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................GAAAGAAAAGTATGAGGCTGA... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........TGTCCGAAGGGGTGGAGCAGTTTGA............................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................................................................................................CAACGCGAACCAGAAAGAAAAGT............. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................TTGACCAGGGAAGCA.................................................................................................................................................................................... | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| TCAAGAAGGTGTCCGAAGGGGTGGAGCAGTTTGAAGATATTTGGCAAAAGGTAAGGACCTAGGAACCTTAAATTTTGTTGTCTGTAGGGCAAAGAATCAAGAAGAGGTTCAGGGCCTTGGGGTCTTGACCAGGGAAGCATCTCTAAGATCGATTGTGGGGAGGGGGTCCTAGATTTCCCAGCCGAGGAAGAAAGAGACTGGACCTTTACAGCTATGGCGAGTTCTGGGCAGCCACTTTCACATTGACTGCTGAATTCTTCATCCCTCAGCTCCACAATGCAGCCAACGCGAACCAGAAAGAAAAGTATGAGGCTGACCT ....................................................................................((((((............((((((((((..((((((........)))))).....(((((....(((..((.(((((.((....))...)))))))..))).......)))))))))))))))...((....))..))))))............................................................................................. ........................................................................73........................................................................................................................................................227.......................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|