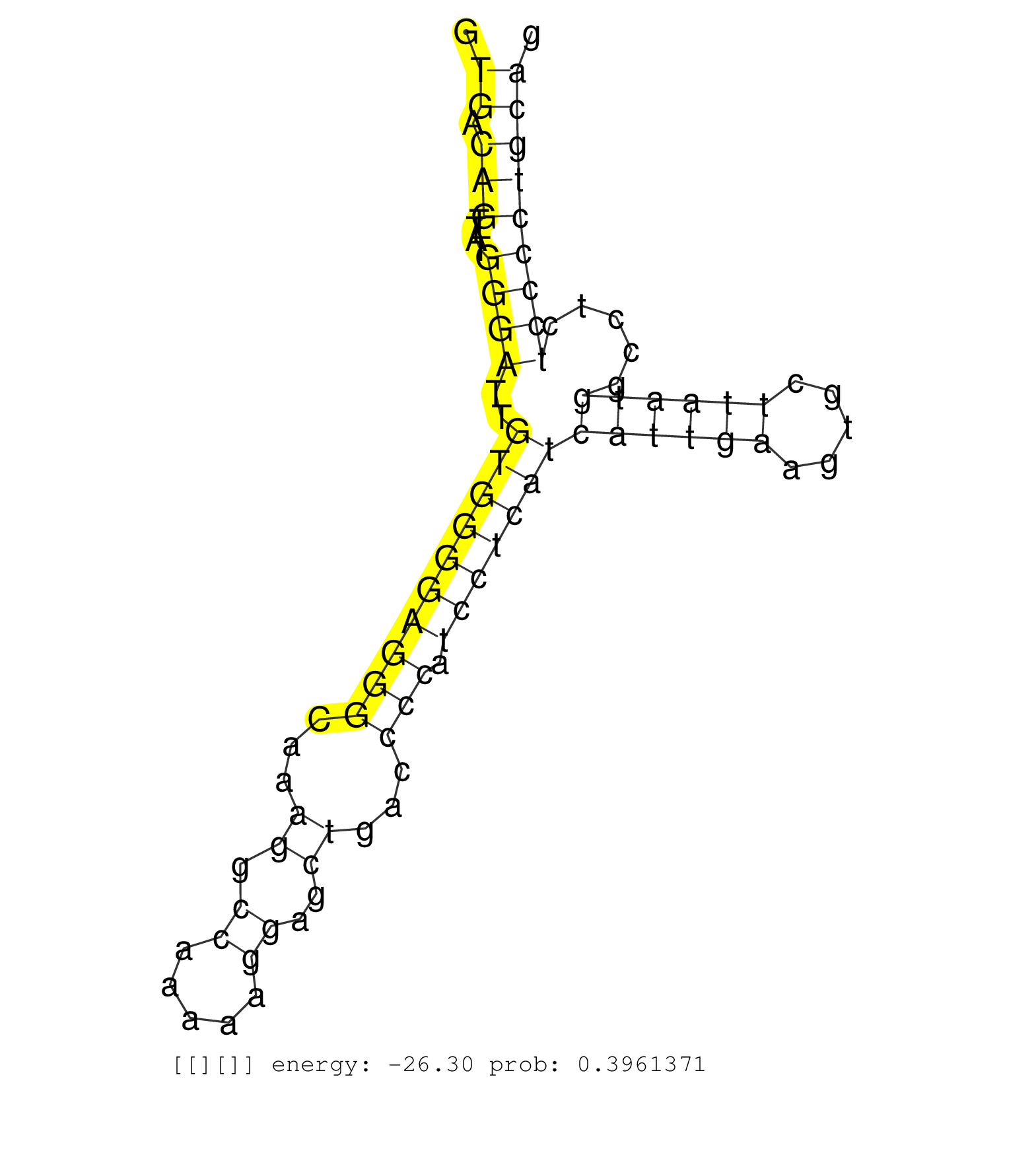
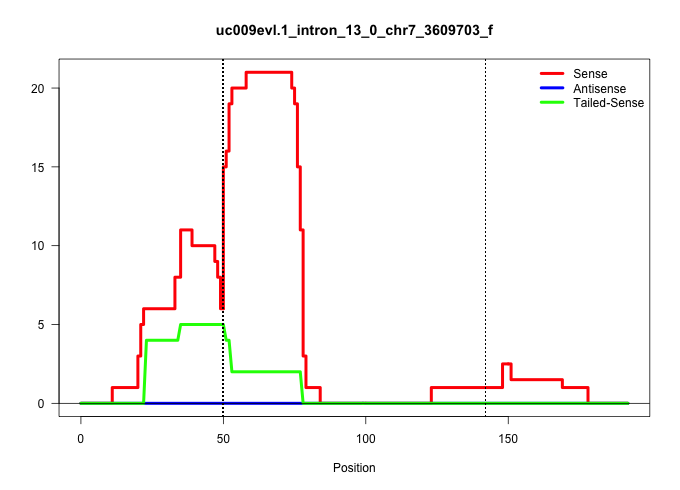
| Gene: Cnot3 | ID: uc009evl.1_intron_13_0_chr7_3609703_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| CAGCTCTGGTATTGAGGACCCTGTGCCAACGTTACACCTAACTGATCGAGGTGACAGTTATGGGATTGTGGGGAGGGCAAAGGCCAAAAAGGAGCTGACCCCATCCTCATCATTGAAGTGCTTAATGGCCTCTCCCCTGCAGACATCATCCTGAGCAGCACATCAGCACCACCCACCTCATCCCAGCCACCC ...................................................((.(((....((((..((((((((((...((.((.....))..))...))).)))))))((((((.....)))))).....)))))))))................................................... ..................................................51.........................................................................................142................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGACAGTTATGGGATTGTGGGGAGGGC.................................................................................................................. | 28 | 1 | 16.00 | 16.00 | 11.00 | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - |
| ..................................................GTGACAGTTATGGGATTGTGGGGAGG.................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................ACCTAACTGATCGAGGT............................................................................................................................................ | 17 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................GACAGTTATGGGATTGTGGGGAGGGC.................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................ACACCTAACTGATCGAG.............................................................................................................................................. | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................GACAGTTATGGGATTGTGGGGAGGGCA................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| ....................................................GACAGTTATGGGATTGTGGGGAGGG................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGACAGTTATGGGATTGTGGGGAGGG................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGCCAACGTTACACCTAACTGATCtag.............................................................................................................................................. | 27 | tag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........TTGAGGACCCTGTGCCAACGTTACACCT......................................................................................................................................................... | 28 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................TGTGCCAACGTTACACCTAACTGATCGAG.............................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCCTGAGCAGCACATCAGCACCACCCACCT.............. | 30 | 2 | 1.00 | 1.00 | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TATGGGATTGTGGGGAGGGCAAAGGC............................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGACAGTTATGGGATTGTGGGGAG..................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................GACAGTTATGGGATTGTGGGGAGGac.................................................................................................................. | 26 | ac | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................CTGTGCCAACGTTACACCTAACTGATCG................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................GTGCCAACGTTACACCTAACTGATCGA............................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGCCAACGTTACACCTAACTGATCGAGa............................................................................................................................................. | 28 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGACAGTTATGGGATTGTGGGGAGGtt.................................................................................................................. | 28 | tt | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................CTGTGCCAACGTTACACCTAACTGATC................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGACAGTTATGGGATTGTGGGGAGGG................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGCCAACGTTACACCTAACTGATCGAGac............................................................................................................................................ | 29 | ac | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AATGGCCTCTCCCCTGCAGACATCATCC......................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGACAGTTATGGGATTGTGGGGA...................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................ACCTAACTGATCGAGaca........................................................................................................................................... | 18 | aca | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................ACAGTTATGGGATTGTGGGGAGGGC.................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................TGCCAACGTTACACCTAACTGATCGAGaca........................................................................................................................................... | 30 | aca | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TGTGCCAACGTTACACCTAACTGATCGA............................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCCTGAGCAGCACATCAGCAC....................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| CAGCTCTGGTATTGAGGACCCTGTGCCAACGTTACACCTAACTGATCGAGGTGACAGTTATGGGATTGTGGGGAGGGCAAAGGCCAAAAAGGAGCTGACCCCATCCTCATCATTGAAGTGCTTAATGGCCTCTCCCCTGCAGACATCATCCTGAGCAGCACATCAGCACCACCCACCTCATCCCAGCCACCC ...................................................((.(((....((((..((((((((((...((.((.....))..))...))).)))))))((((((.....)))))).....)))))))))................................................... ..................................................51.........................................................................................142................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|