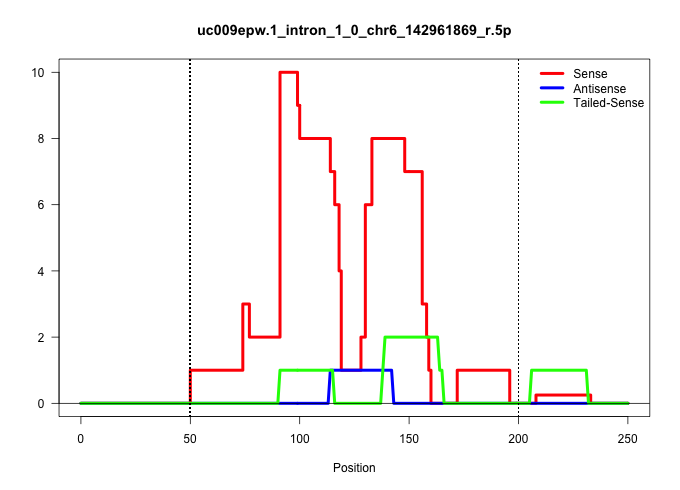
| Gene: 5730419I09Rik | ID: uc009epw.1_intron_1_0_chr6_142961869_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(13) TESTES |
| TCGGCATCATTAACATGTTTTTCATTCGGGAAACCACTTCTCTCCGCGAGGTAACTTACAAAACGGAAGGTTGTTTAAGCCCATTTGTTTGTAGAAGTTGTAGATCCTTGTTGTAAAACACATAATTTTCTCCTGTATTTGAAACTGCACTGCCGGGACACGTGCAGGTTTCTAATGAGGTGGTTTGGGTTCTTTTTTTTTTTTTTTCTTTTTGGTTTTTGAGACAGGCTTTCTCTGTGTAGCCCTGGCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................TCCTGTATTTGAAACTGCACTGCCGG.............................................................................................. | 26 | 1 | 4.00 | 4.00 | - | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................TAGAAGTTGTAGATCCTTGTTGTAAAAC................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TAGAAGTTGTAGATCCTTGTTGTAAAA.................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TAGAAGTTGTAGATCCTTGTTGTAt...................................................................................................................................... | 25 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGAAACTGCACTGCCGGGACACGTaca.................................................................................... | 27 | aca | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................TAGAAGTTGTAGATCCTTGTTGTAA...................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TAGAAGTTGTAGATCCTTGTTGT........................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................TGTATTTGAAACTGCACTGCCGGGAC........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TCTCCTGTATTTGAAACTGCACTGCCGGGA............................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................TTAAGCCCATTTGTTTGTAGAAGTT....................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................CATAATTTTCTCCTGTATTTGAAACTGC...................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................TCTTTTTGGTTTTTGAGACAGGCTTa.................. | 26 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGTATTTGAAACTGCACTGCCGGGACA.......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TAGAAGTTGTAGATCCTTGTTGTAAAACA.................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAACTTACAAAACGGAAGGTTGTTTA............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TAATGAGGTGGTTTGGGTTCTTTT...................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................TTAAGCCCATTTGTTTGTAGAAGTTG...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTGAAACTGCACTGCCGGGACACGTa...................................................................................... | 26 | a | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TTTTTGGTTTTTGAGACAGGCTTTC................. | 25 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| TCGGCATCATTAACATGTTTTTCATTCGGGAAACCACTTCTCTCCGCGAGGTAACTTACAAAACGGAAGGTTGTTTAAGCCCATTTGTTTGTAGAAGTTGTAGATCCTTGTTGTAAAACACATAATTTTCTCCTGTATTTGAAACTGCACTGCCGGGACACGTGCAGGTTTCTAATGAGGTGGTTTGGGTTCTTTTTTTTTTTTTTTCTTTTTGGTTTTTGAGACAGGCTTTCTCTGTGTAGCCCTGGCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................AAAACACATAATTTTCTCCTGTATTTGAA........................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |