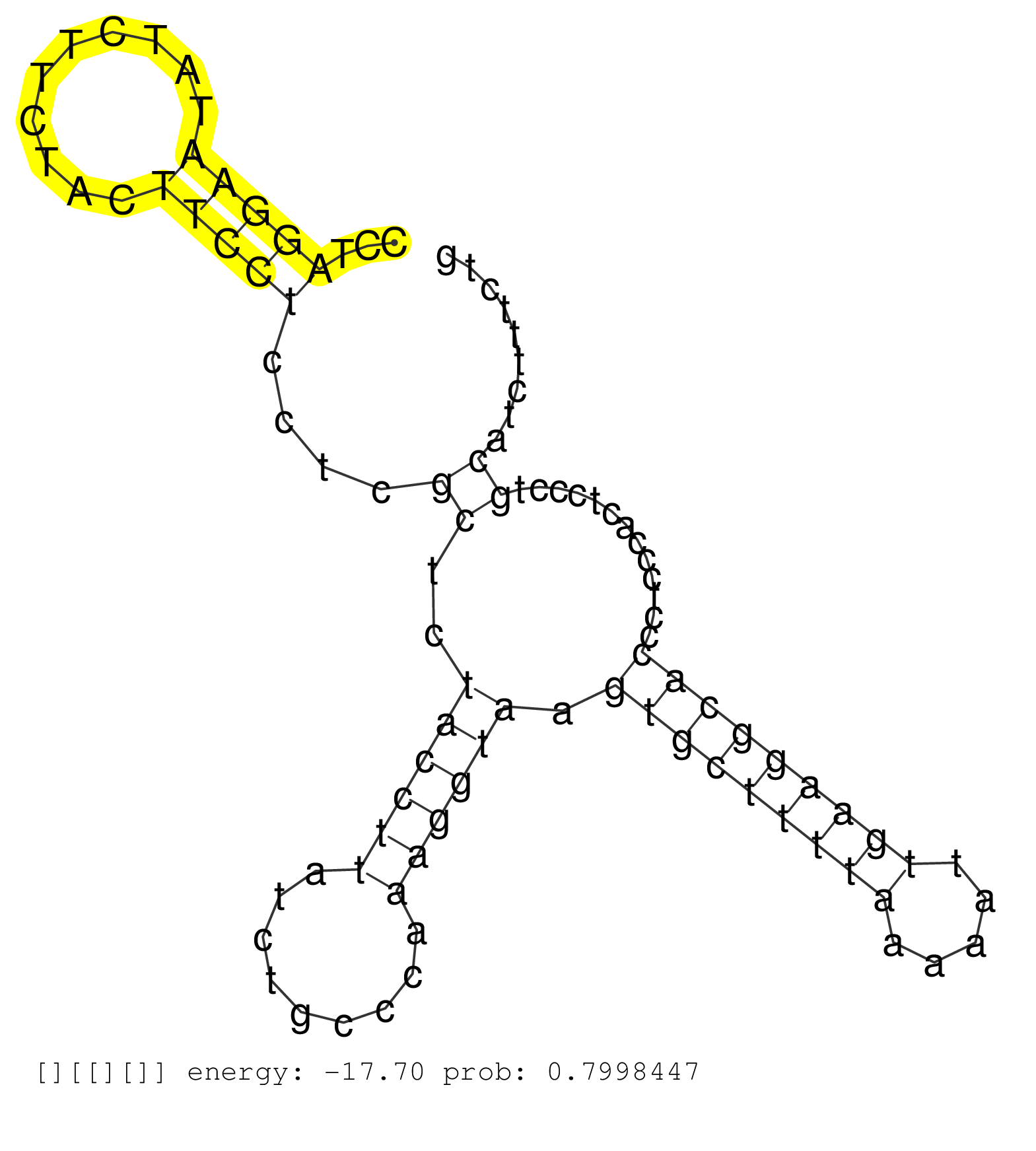
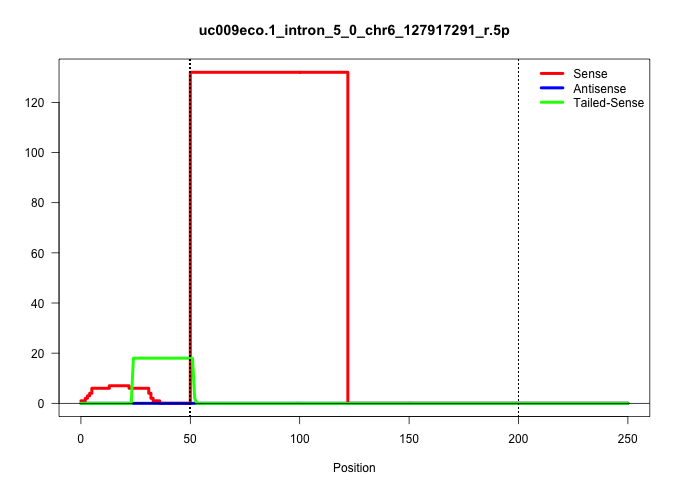
| Gene: Tspan9 | ID: uc009eco.1_intron_5_0_chr6_127917291_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(10) TESTES |
| GTCTCTGCTGCTTGAAGTACACGATGTTCCTCTTCAATTTGATATTCTGGGTAAGTCCTTACTGTTGAGTCTTCCGTTGCTCCTTCTCCTTGCTTTCCACCCTAGGAATATCTTCTACTTCCTCCTCGCTCTACCTTATCTGCCCAAAGGTAAGTGCTTTTAAAAATTGAAGGCACCCTCCCACTCCCTGCATCTTTCTGTTCCCTTTTCCCTCTTCTCCATGGCTGAATGGAGGCTTCAACTTGTCAGT .......................................................................................................(((((..........)))))....((..((((((.........)))))).(((((((((.....))))))))).............))........................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTCCTTACTGTTGAGTCTTCCGTTGCTCCTTCTCCTTGCTTTCCACCC.................................................................................................................................................... | 52 | 1 | 139.00 | 139.00 | 139.00 | - | - | - | - | - | - | - | - | - |
| ........................TGTTCCTCTTCAATTTGATATTCTGGct...................................................................................................................................................................................................... | 28 | ct | 16.00 | 0.00 | - | 1.00 | 5.00 | 2.00 | 4.00 | 1.00 | 3.00 | - | - | - |
| ........................TGTTCCTCTTCAATTTGATATTCTGGctc..................................................................................................................................................................................................... | 29 | ctc | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - |
| .....TGCTGCTTGAAGTACACGATGTTCCTCT......................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .....TGCTGCTTGAAGTACACGATGTTCCTC.......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ...TCTGCTGCTTGAAGTACACGATGTTCCTC.......................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ..CTCTGCTGCTTGAAGTACACGATGTTCCT........................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .............GAAGTACACGATGTTCCTCTTCA...................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ....CTGCTGCTTGAAGTACACGATGTTCCT........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| GTCTCTGCTGCTTGAAGTACACGATGTTCCTCTTCAATTTGATATTCTGGGTAAGTCCTTACTGTTGAGTCTTCCGTTGCTCCTTCTCCTTGCTTTCCACCCTAGGAATATCTTCTACTTCCTCCTCGCTCTACCTTATCTGCCCAAAGGTAAGTGCTTTTAAAAATTGAAGGCACCCTCCCACTCCCTGCATCTTTCTGTTCCCTTTTCCCTCTTCTCCATGGCTGAATGGAGGCTTCAACTTGTCAGT .......................................................................................................(((((..........)))))....((..((((((.........)))))).(((((((((.....))))))))).............))........................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................CATCTTTCTGTTCCCTTTaggg.......................................... | 22 | aggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................GCATCTTTCTGTTCCCTTTTCCCTC.................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |