
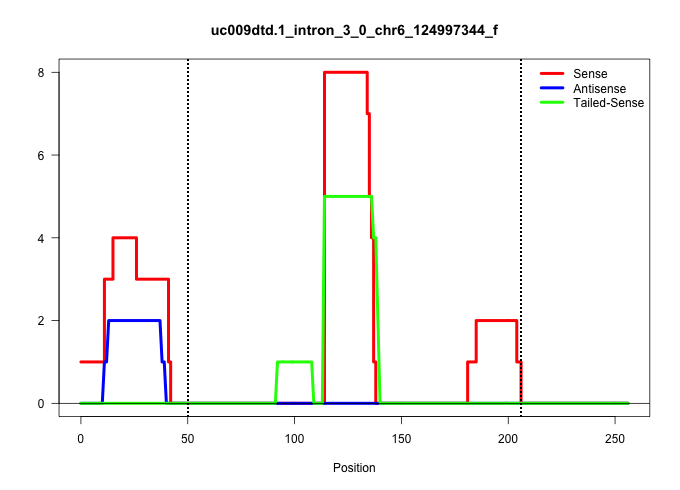
| Gene: Ing4 | ID: uc009dtd.1_intron_3_0_chr6_124997344_f | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| ACAGATCGAGTCCAGTGACTATGACAGCTCTTCTAGCAAAGGCAAAAAGAGTGAGGAGGGGCGGGGCTGTGAGATGCGGGCAAGGAGTCAGAGGGAGAAGAGGCCAACTCCAGCTAGGAGGGAGCGGCACTGCCTGGACCAGGAGCAAAGAGAACTTAACCCATCTTTTTGTCTCTGCCTCCCGGCTTACCCTTTCTTCCTAGAAGGCCGGACCCAAAAGGAGAAAAAAGCTGCCAGAGCCCGTTCCAAAGGGAAA .................................................................................................................(((((((((((.((....(((.(((.(((..(((((((((..........)))))))))..)))..))).)))...)).)))))))))))..................................................... .............................................................................................................110.............................................................................................206................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................TAGGAGGGAGCGGCACTGCCTGG....................................................................................................................... | 23 | 1 | 3.00 | 3.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CACTGCCTGGACCAGGAGCAAAGA......................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TAGGAGGGAGCGGCACTGCCTGGAaa.................................................................................................................... | 26 | aa | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TAGGAGGGAGCGGCACTGCCT......................................................................................................................... | 21 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TAGGAGGGAGCGGCACTGCCTGGA...................................................................................................................... | 24 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........CCAGTGACTATGACAGCTCTTCTAGCAAAG....................................................................................................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TAGGAGGGAGCGGCACTGCCTGGAt..................................................................................................................... | 25 | t | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................TAGGAGGGAGCGGCACTGCCTGa....................................................................................................................... | 23 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............TGACTATGACAGCTCTTCTAGCAAAGG...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................TAGGAGGGAGCGGCACTGCCTGGAa..................................................................................................................... | 25 | a | 1.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................GGGAGAAGAGGCCAtaa................................................................................................................................................... | 17 | taa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TAGGAGGGAGCGGCACTGCCTG........................................................................................................................ | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CCGGCTTACCCTTTCTTCCTAGAAG.................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................TAGGAGGGAGCGGCACTGCC.......................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................................................CTTACCCTTTCTTCCTAGA.................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CTGGACCAGGAGCAAA........................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| ACAGATCGAGTCCAGTGACTATGACAGCTCTTCTAGCAAAGGCAAAAAGAGTGAGGAGGGGCGGGGCTGTGAGATGCGGGCAAGGAGTCAGAGGGAGAAGAGGCCAACTCCAGCTAGGAGGGAGCGGCACTGCCTGGACCAGGAGCAAAGAGAACTTAACCCATCTTTTTGTCTCTGCCTCCCGGCTTACCCTTTCTTCCTAGAAGGCCGGACCCAAAAGGAGAAAAAAGCTGCCAGAGCCCGTTCCAAAGGGAAA .................................................................................................................(((((((((((.((....(((.(((.(((..(((((((((..........)))))))))..)))..))).)))...)).)))))))))))..................................................... .............................................................................................................110.............................................................................................206................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................GCTGCCAGAGCCCGTTCCAAAGGGAA. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CGGCTTACCCTTTCcttc............................................................ | 18 | cttc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........CCAGTGACTATGACAGCTCTTCTAGCAAA........................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............AGTGACTATGACAGCTCTTCTAGCA.......................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |