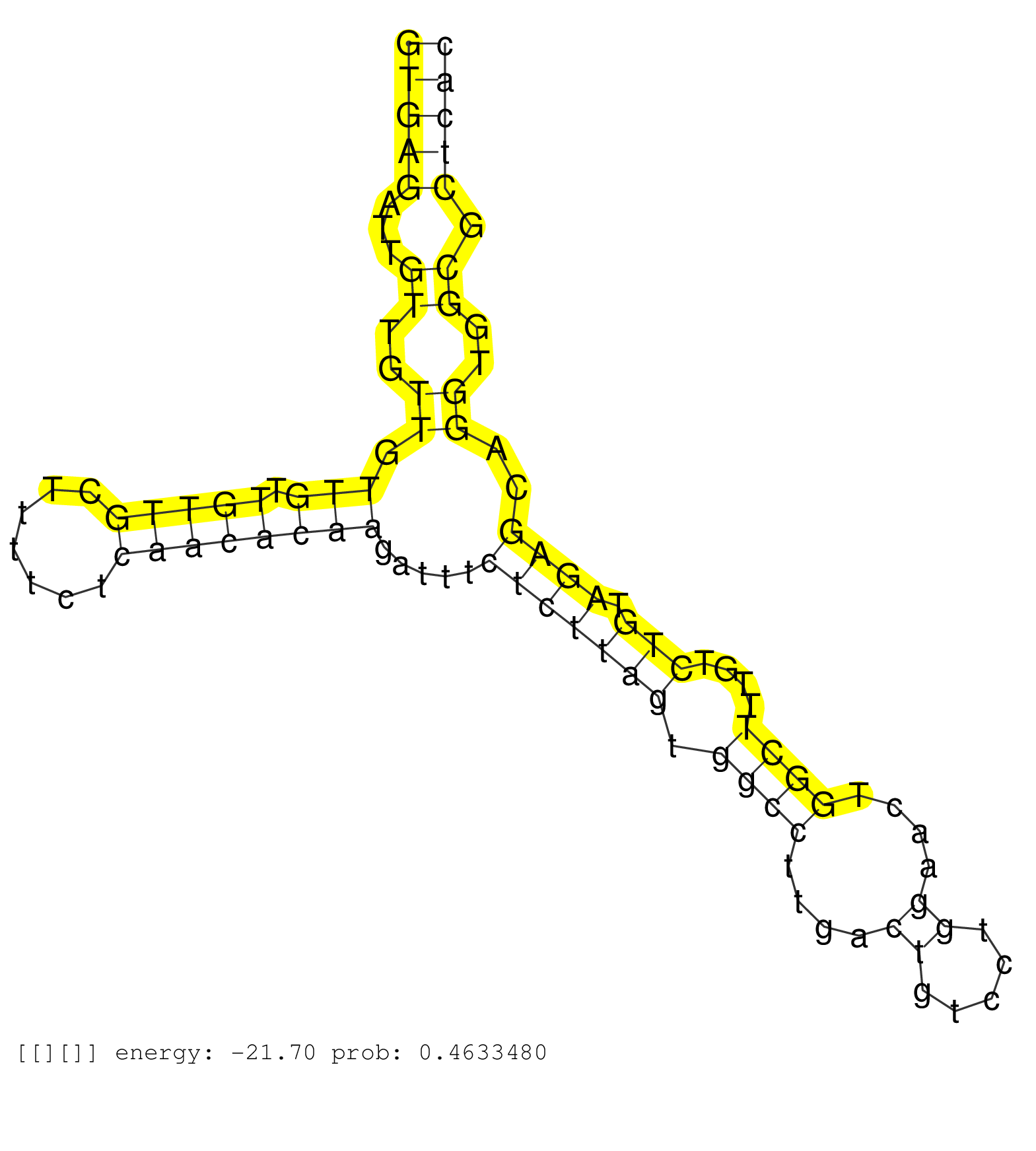

| Gene: Atp6v1e1 | ID: uc009dnr.1_intron_6_0_chr6_120758490_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(20) TESTES |
| CTTTCATTGAACAAGAAGCCAATGAGAAAGCAGAAGAAATAGATGCAAAGGTGAGATTGTTGTTGTTGTTGTTGCTTTTCTCAACACAAGATTTCTCTTAGTGGCCTTGACTGTCCTGGAACTGGCTTTGTCTGTAGAGCAGGTGGCGCTCACAGGGGTGCCTGCCTCCGCCTCCCGGGTGCTGGGACTAGAGGCATGTTTCACCACTGTCCAGGATGTGTCTTGCTTTTATTATGTCATACAAAGAAAC ..................................................(((((...((..((.(((.(((((.......)))))))).....(((((((.((((....((.....))....))))....))).))))..))..)).)))))................................................................................................. ..................................................51....................................................................................................153............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......TGAACAAGAAGCCAATGAGAAAGCAGA........................................................................................................................................................................................................................ | 27 | 1 | 18.00 | 18.00 | 2.00 | 2.00 | 1.00 | 8.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................TGGCTTTGTCTGTAGAGCAGGTGGCGC..................................................................................................... | 27 | 1 | 11.00 | 11.00 | 1.00 | 1.00 | - | - | 2.00 | - | - | 2.00 | 2.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................TGGAACTGGCTTTGTCTGTAGAGCAGG........................................................................................................... | 27 | 1 | 6.00 | 6.00 | 3.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGAACAAGAAGCCAATGAGAAAGCAGAA....................................................................................................................................................................................................................... | 28 | 1 | 5.00 | 5.00 | - | 2.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TTGAACAAGAAGCCAATGAGAAAGCAG......................................................................................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| .......TGAACAAGAAGCCAATGAGAAAGCAGAAG...................................................................................................................................................................................................................... | 29 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAGAAAGCAGAAGAAATAGATGCAAAG........................................................................................................................................................................................................ | 28 | 1 | 4.00 | 4.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................TGGAACTGGCTTTGTCTGTAGAGCAGGT.......................................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGATTGTTGTTGTTGTTGTTGCT.............................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TTGAACAAGAAGCCAATGAGAAAGCAGA........................................................................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGGAACTGGCTTTGTCTGTAGAGCA............................................................................................................. | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGATTGTTGTTGTTGTTGTTGCTT............................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGAACAAGAAGCCAATGAGAAAGCAGAAGAAA................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGATTGTTGTTGTTccc.................................................................................................................................................................................... | 19 | ccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................TTGTCTGTAGAGCAGGTGGCGCTCACA................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGATTGTTGTTGTTGTTGTTGC............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAGAAAGCAGAAGAAATAGATGCAAA......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGGGACTAGAGGCATGTTTCACCACT.......................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TTGAACAAGAAGCCAATGAGAAAGCAGAA....................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........AACAAGAAGCCAATGAGAAAGCAGAAGAAA................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TTCTCTTAGTGGCCTTGACTGTCCTGG................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACTGTCCTGGAACTGGCTTTGTagt.................................................................................................................... | 27 | agt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGGAACTGGCTTTGTCTGTAGAGCttg........................................................................................................... | 27 | ttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................TGGAACTGGCTTTGTCTGTAGAGCAG............................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TAGTGGCCTTGACTGTCCTGGAACTG.............................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGGGACTAGAGGCATGTTTCACCACTGTC....................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAGAAAGCAGAAGAAATAGATGCAAAGGc...................................................................................................................................................................................................... | 30 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................AATGAGAAAGCAGAAGAAATAGATGCAAAGG....................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................TGAGATTGTTGTTGTTGTTGTTGCTT............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGAACAAGAAGCCAATGAGAAAGCAGAAGAAAT.................................................................................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GCTGGGACTAGAGGCATGTTTCACCAC........................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TAGATGCAAAGGTGAGATTGTTGTT.......................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......TTGAACAAGAAGCCAATGAGAAAGC........................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................CCCGGGTGCTGGGACTAGAGGCATGTT.................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......TTGAACAAGAAGCCAATGAGAAAGCAGAAG...................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTTTCATTGAACAAGAAGCCAATGAGAAAGCAGAAGAAATAGATGCAAAGGTGAGATTGTTGTTGTTGTTGTTGCTTTTCTCAACACAAGATTTCTCTTAGTGGCCTTGACTGTCCTGGAACTGGCTTTGTCTGTAGAGCAGGTGGCGCTCACAGGGGTGCCTGCCTCCGCCTCCCGGGTGCTGGGACTAGAGGCATGTTTCACCACTGTCCAGGATGTGTCTTGCTTTTATTATGTCATACAAAGAAAC ..................................................(((((...((..((.(((.(((((.......)))))))).....(((((((.((((....((.....))....))))....))).))))..))..)).)))))................................................................................................. ..................................................51....................................................................................................153............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|