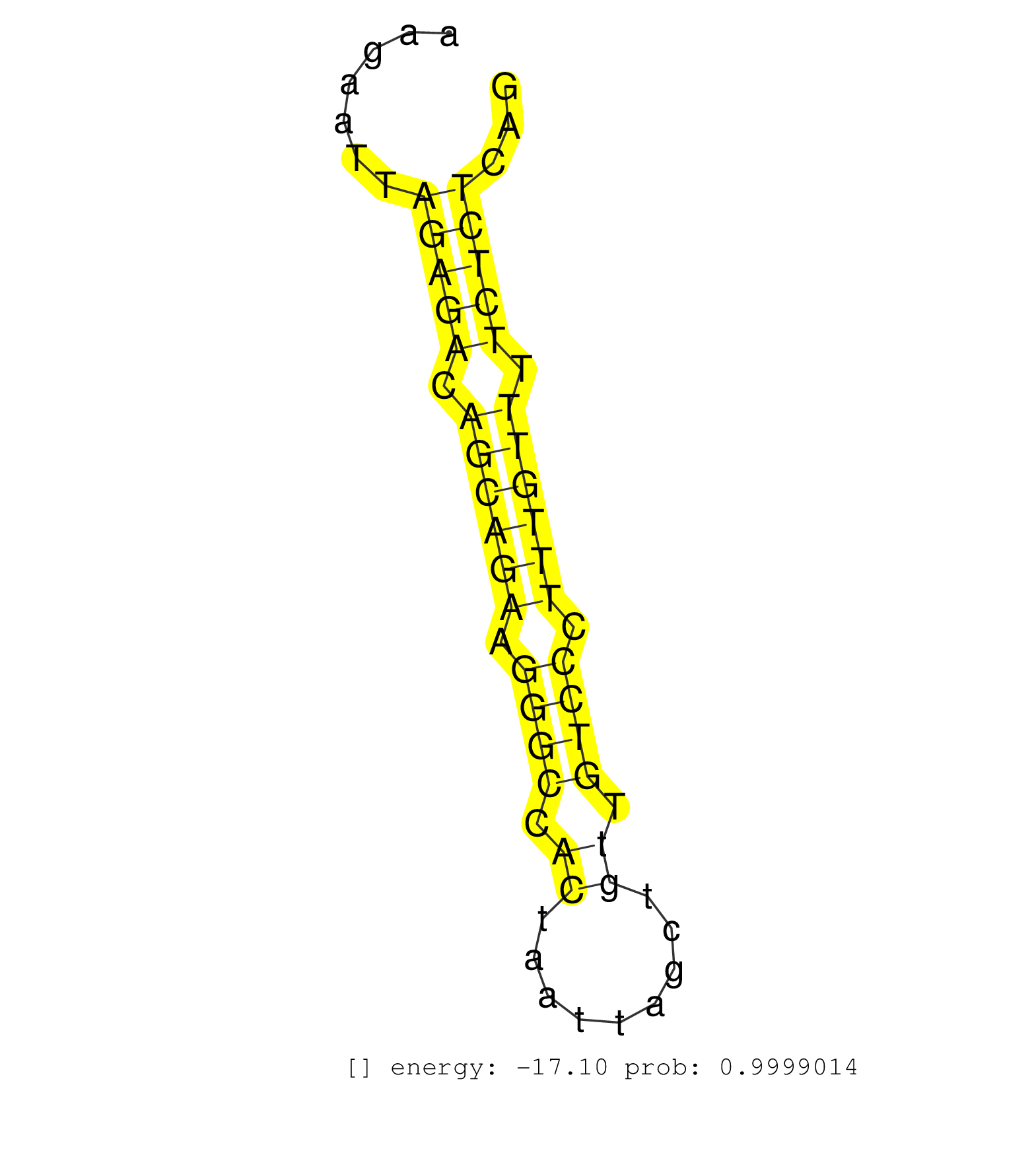

| Gene: Tatdn2 | ID: uc009dhg.1_intron_3_0_chr6_113654901_f.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(22) TESTES |
| TGTCTCCCTAGGGGACCAGTGAGGATTGCAGGTTGGTGAGCAGCACAAGATAGTGATTTGAGGGCTTTGAGGAATCGGCAATGTAGAAAGGACTAGTAGATGGTGATCATGTTTACAATTCTATTGTACAGTTTAGAAGCAAAGAATTAGAGACAGCAGAAGGGCCACTAATTAGCTGTTGTCCCTTTGTTTTCTCTCAGGTGTTTGAGAGGCAGCTGAAGCTGGCTGTGTCTCTGAAGAAGCCCTTGGT ....................................................................................................................................................(((((.((((((.((((.((.........)).)))).)))))).)))))..................................................... .............................................................................................................................................142.......................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................TGTCCCTTTGTTTTCTCTCAG.................................................. | 21 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGTCCCTTTGTTTTCTCTCAGt................................................. | 22 | t | 6.00 | 7.00 | - | 2.00 | - | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTAGAGACAGCAGAAGGGCCAC.................................................................................. | 22 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGAGACAGCAGAAGGGCCAC.................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGAGGAATCGGCAATGTAGAAAGGAC............................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGTCCCTTTGTTTTCTCTCAGa................................................. | 22 | a | 2.00 | 7.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................CAGTGAGGATTGCAGGT......................................................................................................................................................................................................................... | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGAGACAGCAGAAGGGCCA................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTAGAGACAGCAGAAGGGCCAaat................................................................................ | 24 | aat | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TAGAAGCAAAGAATTAGAGACAGCAGA.......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGAGACAGCAGAAGGGCCACT................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................TTAGAGACAGCAGAAGGGCCACT................................................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTAGAGACAGCAGAAGGGCCACat................................................................................ | 24 | at | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAAGCTGGCTGTGTCTCTGAAGAAGCC...... | 28 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ATTAGAGACAGCAGAAGGGCC.................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................TTAGAGACAGCAGAAGGGCCAat................................................................................. | 23 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................TGATTTGAGGGCTTTGAGGAATCGG............................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAAGCTGGCTGTGTCTCTGAAGAAGacc..... | 29 | acc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................AAAGGACTAGTAGATG.................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................TTAGAGACAGCAGAAGGGCCACTAt............................................................................... | 25 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTAGAGACAGCAGAAGGGCCAt.................................................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................TTAGAGACAGCAGAAGGGCCACTt................................................................................ | 24 | t | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAAGCTGGCTGTGTCTCTGAAGAAGC....... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................CTGAAGCTGGCTGTGTCTCTGAAG........... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGGCTGTGTCTCTGAAGAAGCC...... | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................GCTGTGTCTCTGAAGAAGCCCTTGG. | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ............................................................................................................................................................................................................................GCTGGCTGTGTCTCTGAAGAAGCCCT.... | 26 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................CTGTGTCTCTGAAGAAGCCC..... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| TGTCTCCCTAGGGGACCAGTGAGGATTGCAGGTTGGTGAGCAGCACAAGATAGTGATTTGAGGGCTTTGAGGAATCGGCAATGTAGAAAGGACTAGTAGATGGTGATCATGTTTACAATTCTATTGTACAGTTTAGAAGCAAAGAATTAGAGACAGCAGAAGGGCCACTAATTAGCTGTTGTCCCTTTGTTTTCTCTCAGGTGTTTGAGAGGCAGCTGAAGCTGGCTGTGTCTCTGAAGAAGCCCTTGGT ....................................................................................................................................................(((((.((((((.((((.((.........)).)))).)))))).)))))..................................................... .............................................................................................................................................142.......................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) |
|---|