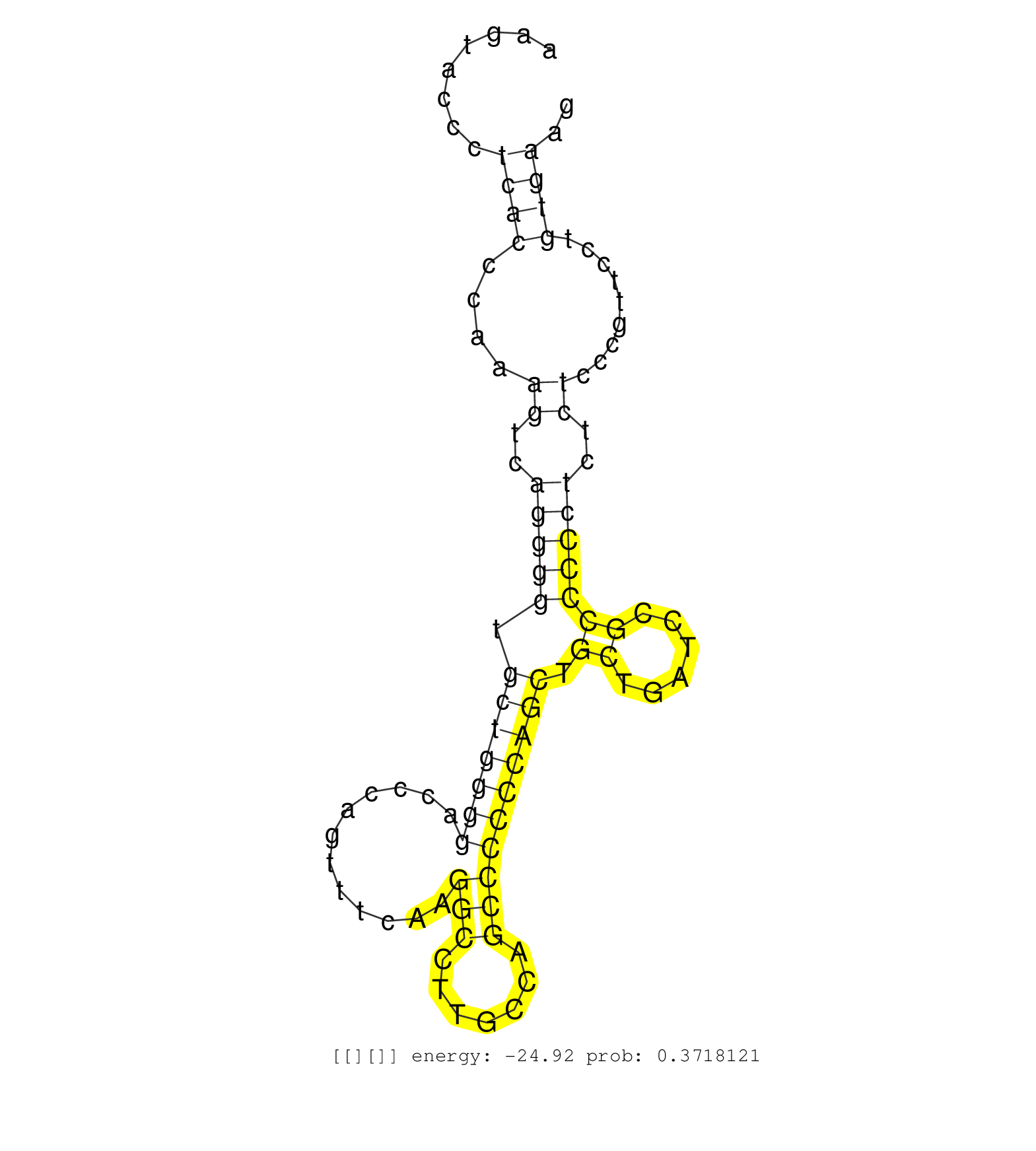
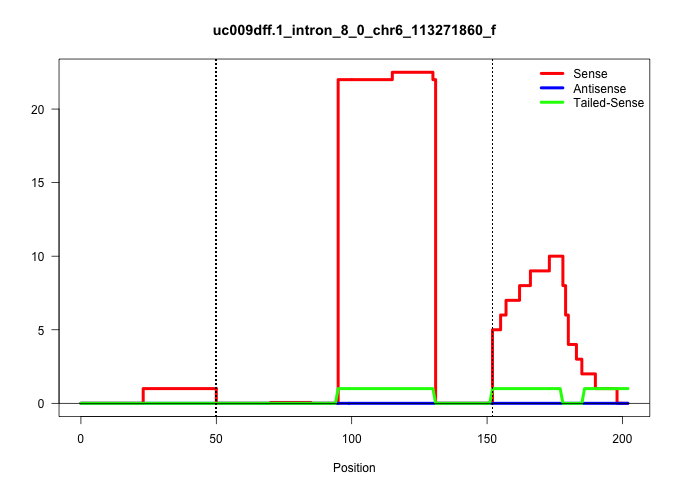
| Gene: Brpf1 | ID: uc009dff.1_intron_8_0_chr6_113271860_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(15) TESTES |
| TGAGAATGAGGCCTACTCCGTGGGCACTGGCCGCGGCGTGGGCCACAGCAGTAAGTACCCTCACCCAAAGTCAGGGGTGCTGGGGACCCAGTTTCAAGGCCTTGCCAGCCCCCCAGCTGCTGATCCGCCCCCTCTCTCCCGTTCCTGTGAAGTGGTAAGAAAGAGTCTGGGTCGAGGAGCTGGCTGGCTGTCAGAGGATGAG ............................................................((((....((..(((((.(((((((............(((.......)))))))))).((......)))))))..)).........)))).................................................... ....................................................53.................................................................................................152................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................AAGGCCTTGCCAGCCCCCCAGCTGCTGATCCGCCCCCTCTCTCCCGTTCCTG....................................................... | 52 | 1 | 121.00 | 121.00 | 121.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................CTGGCTGTCAGAGGATG.. | 17 | 2 | 2.50 | 2.50 | - | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................AGAAAGAGTCTGGGTCGAGGAGCTGGCT................. | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................TGGTAAGAAAGAGTCTGGGTCGAGGAG....................... | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TGGTAAGAAAGAGTCTGGGTCGAGGA........................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - |
| .....................................................................................................................................................................TCTGGGTCGAGGAGCTGGCTGG............... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TAAGAAAGAGTCTGGGTCGAGGAGC...................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................CTGGGTCGAGGAGCTGGCTGGCTG............ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................GAGTCTGGGTCGAGGAGCTGG................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TGGTAAGAAAGAGTCTGGGTCGAGGt........................ | 26 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................GAGGAGCTGGCTGGCTGTCAGAGGA.... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................TGGTAAGAAAGAGTCTGGGTCGAGGAGC...................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................GCACTGGCCGCGGCGTGGGCCACAGCA........................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................................GCTGTCAGAGGATGAtaac | 19 | taac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................GCTGCTGATCCGCCC........................................................................ | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ......................................................................TCAGGGGTGCTGGGG..................................................................................................................... | 15 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - |
| TGAGAATGAGGCCTACTCCGTGGGCACTGGCCGCGGCGTGGGCCACAGCAGTAAGTACCCTCACCCAAAGTCAGGGGTGCTGGGGACCCAGTTTCAAGGCCTTGCCAGCCCCCCAGCTGCTGATCCGCCCCCTCTCTCCCGTTCCTGTGAAGTGGTAAGAAAGAGTCTGGGTCGAGGAGCTGGCTGGCTGTCAGAGGATGAG ............................................................((((....((..(((((.(((((((............(((.......)))))))))).((......)))))))..)).........)))).................................................... ....................................................53.................................................................................................152................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................ATCCGCCCCCTCaaa.................................................................... | 15 | aaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |