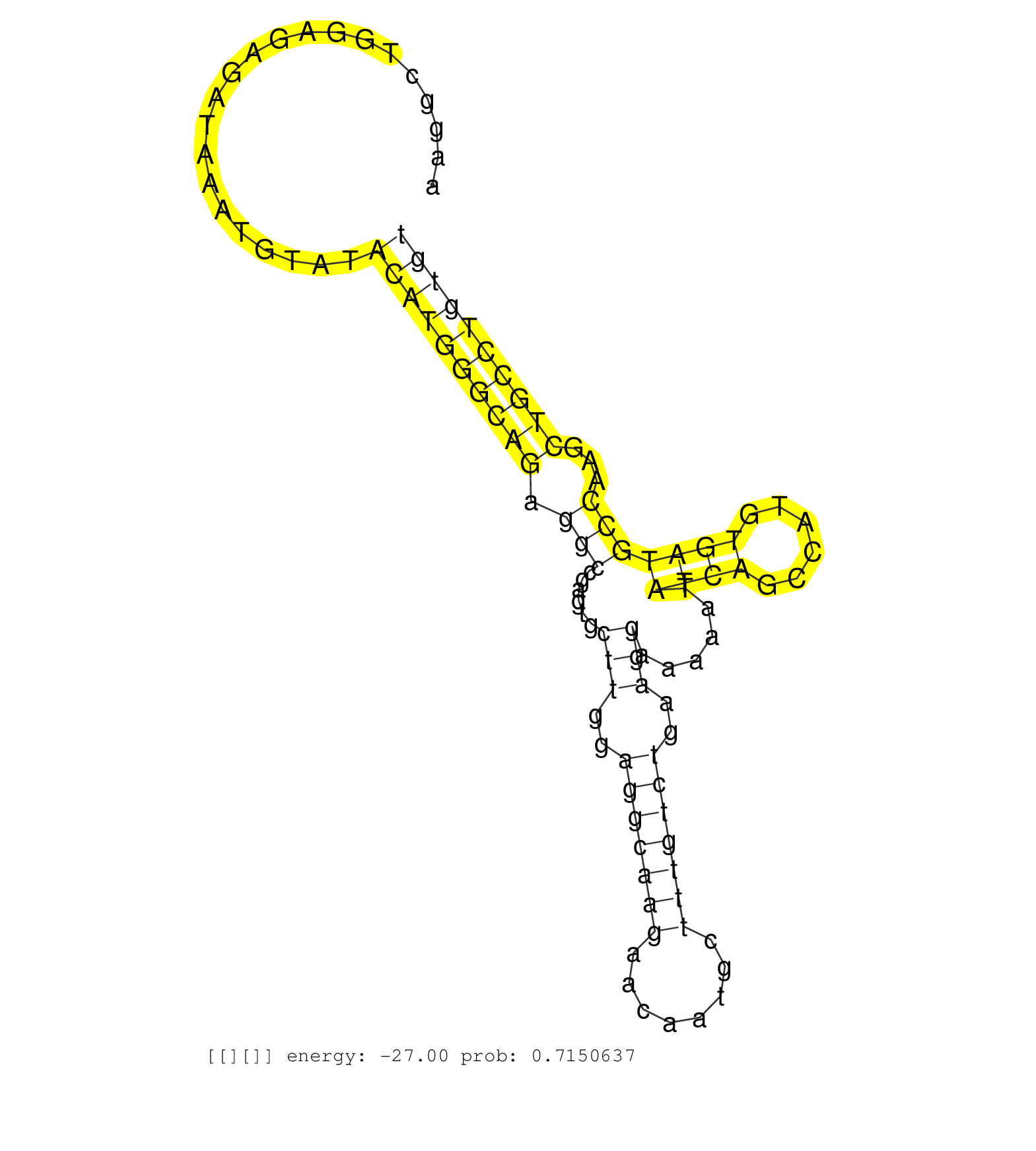
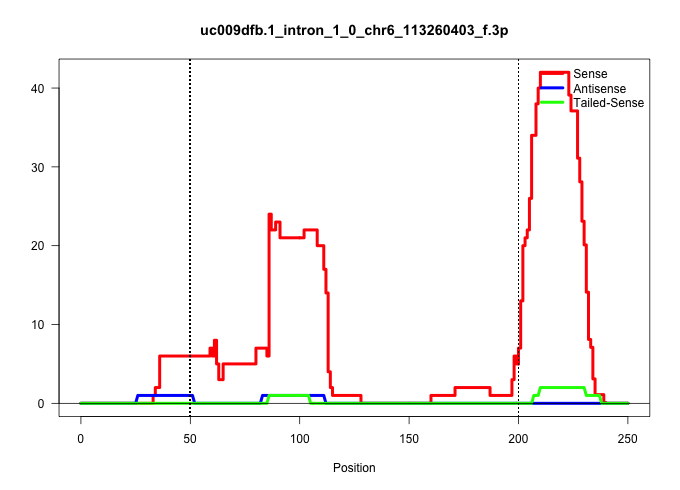
| Gene: Brpf1 | ID: uc009dfb.1_intron_1_0_chr6_113260403_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| GGAAGTTAACTTCTCCCACATTTAGGTCTCCTTTTCTGCAAGACAGGAATGATTCTAATACTATAAATGTGGGGTTAAGGTAAGGCTGGAGAGATAAATGTATACATGGGCAGAGGCCGAGTGCTTGGAGGCAAGAACAATGCTTTGTCTGAAGGAAAAATATCAGCCATGTGATGCCAAGCTGCCTGTGTGCACTGCAGGTACATCGAGAAATCTGCAGAGGAGCTGGATGAGGAGGTGGAGTATGACA .......................................................................................................((((((((((.(((......(((..(((((((........)))))))..)))......((((......)))))))...))))))))))........................................................... .................................................................................82...........................................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................TGGAGAGATAAATGTATACATGGGCAG......................................................................................................................................... | 27 | 1 | 10.00 | 10.00 | 5.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................................CAGGTACATCGAGAAATCTGCAGAGGAGCT....................... | 30 | 1 | 7.00 | 7.00 | - | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ACATCGAGAAATCTGCAGAGGAGCTGG..................... | 27 | 1 | 5.00 | 5.00 | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TACATCGAGAAATCTGCAGAGGAGCT....................... | 26 | 1 | 5.00 | 5.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CGAGAAATCTGCAGAGGAGCTGGAT................... | 25 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................................................................CGAGAAATCTGCAGAGGAGCTGGATG.................. | 26 | 1 | 4.00 | 4.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AGAAATCTGCAGAGGAGCTGGATGAG................ | 26 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TGGAGAGATAAATGTATACATGGGCA.......................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TGGAGAGATAAATGTATACATGGGC........................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCGAGAAATCTGCAGAGGAGCTGGAT................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGTACATCGAGAAATCTGCAGAGGA.......................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................TAAGGCTGGAGAGATAAATGTATACATG.............................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AAATCTGCAGAGGAGCTGGATGAGG............... | 25 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AATGTGGGGTTAAGGTAAGGCTGGAG............................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................TGGAGAGATAAATGTATACATGGGCAGA........................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TATAAATGTGGGGTTAAGGTAAGGCT................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TGCAAGACAGGAATGATTCTAATACTA........................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGTACATCGAGAAATCTGCAGAGG........................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCGAGAAATCTGCAGAGGAGCTGGATG.................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................CATCGAGAAATCTGCAGAGGAGCTGGA.................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TATCAGCCATGTGATGCCAAGCTGCCT............................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................................................................TACATCGAGAAATCTGCAGAGGAGCTG...................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGTACATCGAGAAATCTGCAGAGGA.......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................................................GAAATCTGCAGAGGAGCTGGATGAGGAGGT........... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TTCTGCAAGACAGGAATGATTCTAATACT............................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GTACATCGAGAAATCTGCAGAGGAGCTG...................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ACATCGAGAAATCTGCAGAGGAGCTG...................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GAGAAATCTGCAGAGGAGCTGtat................... | 24 | tat | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGTACATCGAGAAATCTGCAGAGG........................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TGCAAGACAGGAATGATTCTAATA.............................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AAATCTGCAGAGGAGCTGGATGAGGAGt............ | 28 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................ACTATAAATGTGGGGTTAAGGTAAGG..................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GAAATCTGCAGAGGAGCTGGATGAG................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TGCAAGACAGGAATGATTCTAATACT............................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGATGCCAAGCTGCCTGTGTGCACTGCA................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................AGAAATCTGCAGAGGAGCTGGATGA................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................ATCGAGAAATCTGCAGAGGAGCTGGA.................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TCTGCAAGACAGGAATGATTCTAATACT............................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TACATGGGCAGAGGCCGAGTGCTTGG.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GTACATCGAGAAATCTGCAGAGGAGCT....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ACATCGAGAAATCTGCAGAGGAGCTGGA.................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AGAGATAAATGTATACATGGGCAGAG....................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TGGAGAGATAAATGTAgga................................................................................................................................................. | 19 | gga | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................AGCTGGATGAGGAGGTG.......... | 17 | 10 | 0.10 | 0.10 | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGAAGTTAACTTCTCCCACATTTAGGTCTCCTTTTCTGCAAGACAGGAATGATTCTAATACTATAAATGTGGGGTTAAGGTAAGGCTGGAGAGATAAATGTATACATGGGCAGAGGCCGAGTGCTTGGAGGCAAGAACAATGCTTTGTCTGAAGGAAAAATATCAGCCATGTGATGCCAAGCTGCCTGTGTGCACTGCAGGTACATCGAGAAATCTGCAGAGGAGCTGGATGAGGAGGTGGAGTATGACA .......................................................................................................((((((((((.(((......(((..(((((((........)))))))..)))......((((......)))))))...))))))))))........................................................... .................................................................................82...........................................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................AAGAACAATGCTTTGTCTGAAGGAAAAATA........................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................GGCTGGAGAGATAAATGTATACATGGGCA.......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................TCTCCTTTTCTGCAAGACAGGAATGA...................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |