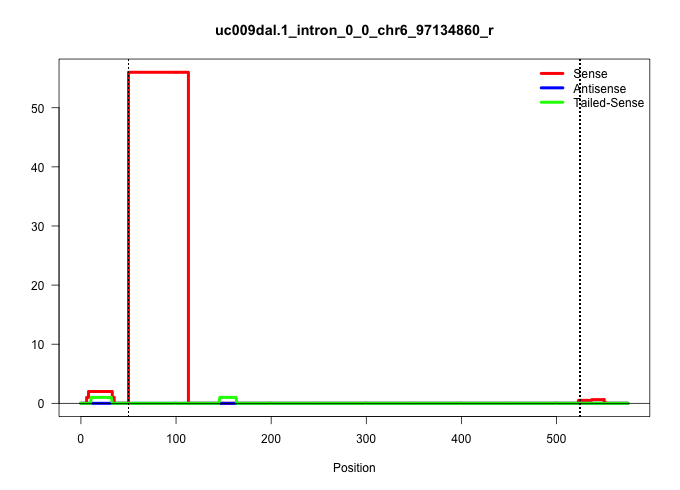
| Gene: Ube1c | ID: uc009dal.1_intron_0_0_chr6_97134860_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) PIWI.ip |
(16) TESTES |
| AACGTCTATTGAAGAACGAACCAGGCCCAATCTTTCCAAAACATTAAAAGGTATTTTAAGTAAATGTGTATAGTTATAGTTTTCTTTTTTAGTGAATGTCTGCTCTCCATCCCTTCTTCCTCCTTCCCTCCTTCCCTCCCCCTCTGTGTGTGTGGGGAAAGGGGGCTGTCTGTCTGTCCGTCCCTGGTACAGGTGCCTTATCCCTGTGCCTGCTGTCTCCTCAGCCCTCAGTGTTTGGAGATAGGATGGATGCCTTTGTGTGGCTTTAGCTAGCCTTGAACTTGCTTATAGCCCAGGCTGGCTTTGAACTCAGATTTATTCATAATTAAATATGACCAAATTGTAATTATTTATATCTCTATAGTAGAATTGATGGCTTCTTTTAATAAAAATAATTTAAAATCCAAGAAATCAAGTTTTAAGACATGTCTGAACTATAAAGCTATAACTTGTGCGTTATGTATGTGTTATTCTTTTTGTTGAGGTATTTATTTTGCTTGGTCTAATGACACTTTTCTATTTTAGAACTGGGACTAGTTGATGGACAAGAACTGGCTGTTGCTGATGTCACTACA .........................................................(((.....(((...(((....((((.((.....)).))))....)))...))).....)))........(((((((...(((((............)))))))))))).......................................................................................................................................................................................................................................................................................................................................................................................................................... ..................................................51...................................................................................................................168..................................................................................................................................................................................................................................................................................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATTTTAAGTAAATGTGTATAGTTATAGTTTTCTTTTTTAGTGAATGTCTG......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 56.00 | 56.00 | 56.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GATGGACAAGAACTGGCTG................. | 19 | 2 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GATGGACAAGAACTGGCTGTTGCTGAT......... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGATGGACAAGAACTGGCTGT................ | 21 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TAGAACTGGGACTAGTTGATGGA.............................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........TTGAAGAACGAACCAGGCCCAATCTTT............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GACTAGTTGATGGACAAGAACTGGCTGTTGCTGA.......... | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........AAGAACGAACCAGGCCCAATat.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | at | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ATGGACAAGAACTGGCTGTTGCTGAT......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................TGTGTGTGGGGAAAcaaa........................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | caaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......TATTGAAGAACGAACCAGGCCCAATCT.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AAGAACTGGCTGTTGCTGATGT....... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GAACTGGGACTAGTTGATGGACAAGAACT...................... | 29 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGAACTGGGACTAGTTGATGGACAAGA......................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTGATGGACAAGAAC....................... | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |
| AACGTCTATTGAAGAACGAACCAGGCCCAATCTTTCCAAAACATTAAAAGGTATTTTAAGTAAATGTGTATAGTTATAGTTTTCTTTTTTAGTGAATGTCTGCTCTCCATCCCTTCTTCCTCCTTCCCTCCTTCCCTCCCCCTCTGTGTGTGTGGGGAAAGGGGGCTGTCTGTCTGTCCGTCCCTGGTACAGGTGCCTTATCCCTGTGCCTGCTGTCTCCTCAGCCCTCAGTGTTTGGAGATAGGATGGATGCCTTTGTGTGGCTTTAGCTAGCCTTGAACTTGCTTATAGCCCAGGCTGGCTTTGAACTCAGATTTATTCATAATTAAATATGACCAAATTGTAATTATTTATATCTCTATAGTAGAATTGATGGCTTCTTTTAATAAAAATAATTTAAAATCCAAGAAATCAAGTTTTAAGACATGTCTGAACTATAAAGCTATAACTTGTGCGTTATGTATGTGTTATTCTTTTTGTTGAGGTATTTATTTTGCTTGGTCTAATGACACTTTTCTATTTTAGAACTGGGACTAGTTGATGGACAAGAACTGGCTGTTGCTGATGTCACTACA .........................................................(((.....(((...(((....((((.((.....)).))))....)))...))).....)))........(((((((...(((((............)))))))))))).......................................................................................................................................................................................................................................................................................................................................................................................................................... ..................................................51...................................................................................................................168..................................................................................................................................................................................................................................................................................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........GAACGAACCAGGccgg...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 16 | ccgg | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................GTGCCTTATCCCTtggc.................................................................................................................................................................................................................................................................................................................................................................................. | 17 | tggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................................................................................................................CAGGCTGGCTTTGAACTCAGAagaa..................................................................................................................................................................................................................................................................... | 25 | agaa | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |