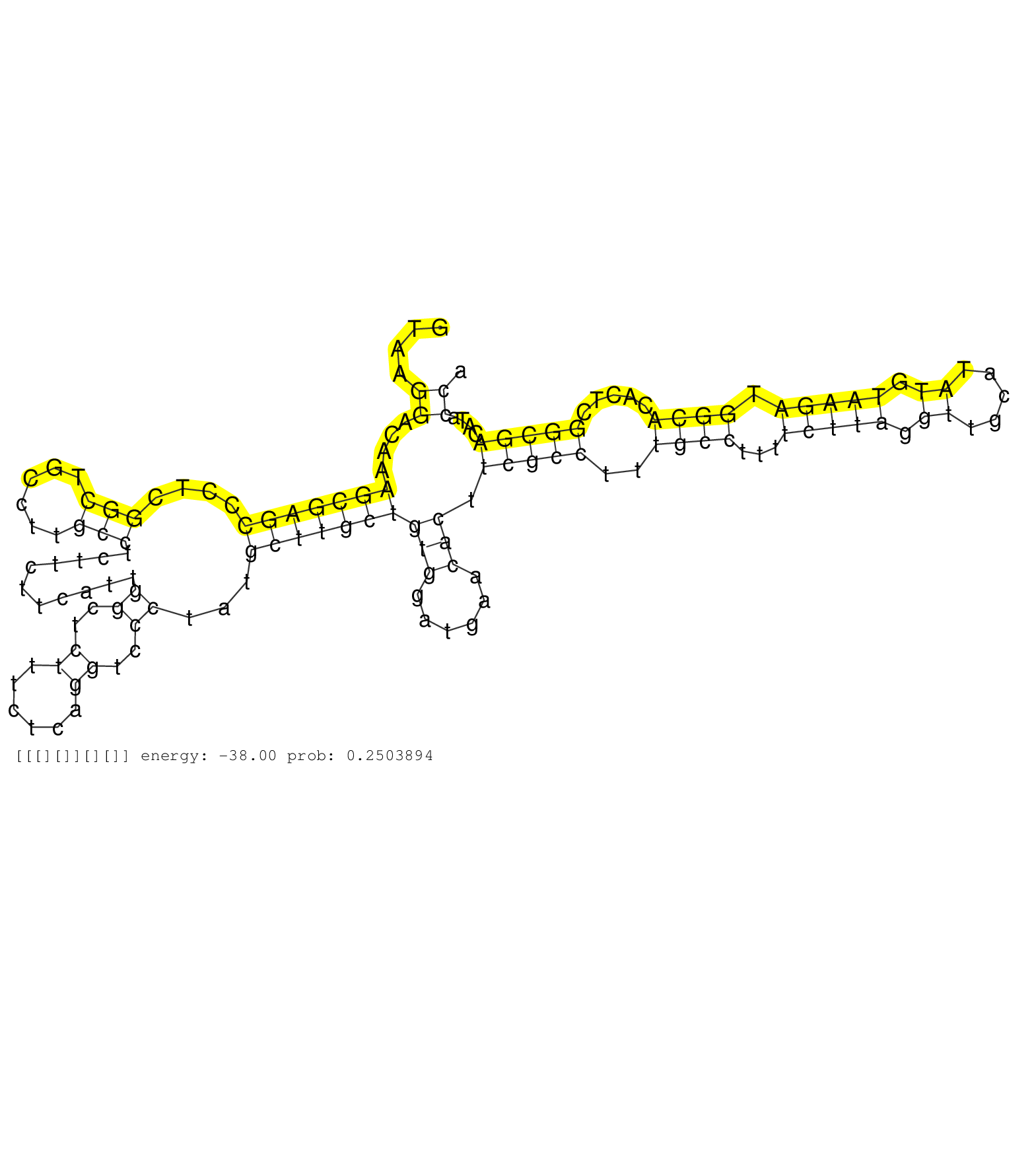
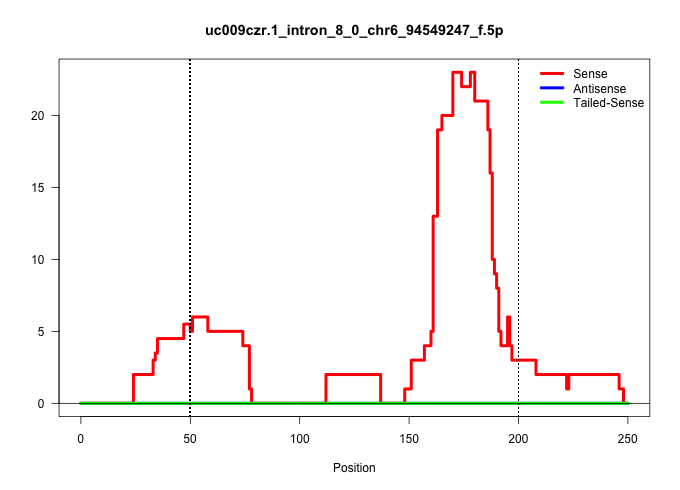
| Gene: Slc25a26 | ID: uc009czr.1_intron_8_0_chr6_94549247_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(12) TESTES |
| AACGTGCTCTCTGCCATGCACGGGGTGTGGCGCTCACAGGGACTAGCCGGGTAAGGACAAAGCGAGCCCTCGGCTGCCTTGCCTCTTCTTCATTGGCTCTTTCTCAGGTCCCTATGCTTGCTGTGGATGAACACTTCGCCTTTGCCTTTTCTTAGGTTGCATATGTAAGATGGCACACTCGGCGACATACCATTCTAGCCACATGGTTCTCGGTCTTGTCCTTGACACTGGATGACATGCAAATGTGTGT ......................................................((....(((((((....(((......)))...........((..((......))..))...)))))))(((......))).(((((..((((...(((((.((.....)).))))).)))).....)))))....))........................................................... ..................................................51...........................................................................................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................TATGTAAGATGGCACACTCGGCGACAT.............................................................. | 27 | 1 | 6.00 | 6.00 | 2.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAAGGACAAAGCGAGCCCTCGGCTGC............................................................................................................................................................................. | 27 | 1 | 6.00 | 6.00 | - | - | - | - | 3.00 | - | - | - | 2.00 | - | 1.00 | - |
| ...................................................................................................................................................................TGTAAGATGGCACACTCGGCGACATACC........................................................... | 28 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TAGCCACATGGTTCTCGGTCTTGTCCT............................ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................TATGCTTGCTGTGGATGAACACTTC................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ........................GTGTGGCGCTCACAGGGACTAGCCGG........................................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TATGTAAGATGGCACACTCGGCGACA............................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................................TGGCACACTCGGCGACATACCATTCT...................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TTAGGTTGCATATGTAAGATGGCACACTC...................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGCATATGTAAGATGGCACACTCGGCGAC................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................ATATGTAAGATGGCACACTCGGCGAC................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGACACTGGATGACATGCAAATGTGT.. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CGGGTAAGGACAAAGCGAGCCCTCGGC................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACAAAGCGAGCCCTCGGCTGa............................................................................................................................................................................. | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................TGTAAGATGGCACACTCGGCGACATA............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................................................TGGCACACTCGGCGACATACCATTCTA..................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTAAGATGGCACACTCGGCGACATAC............................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................ACAGGGACTAGCCGGGTAAGGAC................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................TAAGGACAAAGCGAGCCCTCGGCTGCC............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TAAGATGGCACACTCGGCGACATACCA.......................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCGGCGACATACCATTCTAGCCACATGGTT.......................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GACACTGGATGACATGCAAATGT.... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................TCACAGGGACTAGCCGG........................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................TTCTTAGGTTGCATATGTAAGATGGC............................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTAAGATGGCACACTCGGCGACA............................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................CACAGGGACTAGCCGG........................................................................................................................................................................................................ | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| AACGTGCTCTCTGCCATGCACGGGGTGTGGCGCTCACAGGGACTAGCCGGGTAAGGACAAAGCGAGCCCTCGGCTGCCTTGCCTCTTCTTCATTGGCTCTTTCTCAGGTCCCTATGCTTGCTGTGGATGAACACTTCGCCTTTGCCTTTTCTTAGGTTGCATATGTAAGATGGCACACTCGGCGACATACCATTCTAGCCACATGGTTCTCGGTCTTGTCCTTGACACTGGATGACATGCAAATGTGTGT ......................................................((....(((((((....(((......)))...........((..((......))..))...)))))))(((......))).(((((..((((...(((((.((.....)).))))).)))).....)))))....))........................................................... ..................................................51...........................................................................................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................GACAAAGCGAGCCtag...................................................................................................................................................................................... | 16 | tag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................GACAAAGCGAGCtagg....................................................................................................................................................................................... | 16 | tagg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |