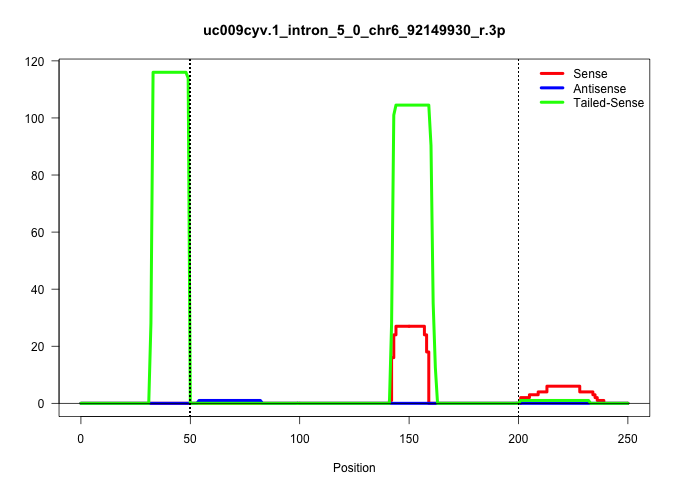
| Gene: Zfyve20 | ID: uc009cyv.1_intron_5_0_chr6_92149930_r.3p | SPECIES: mm9 |
 |
|
(9) OTHER.mut |
(2) OVARY |
(3) PIWI.ip |
(26) TESTES |
| TACCACAAGCTATTTGGACTTGATGGCTTCTTTTCCCACCAGAGTCTCATCCCTGATGCTAGAGTAGGTTCTTCTTGTCCAGAGGTGAAGCCTGCTCTTCCTAGTTCCCAAGTCGCATCTGTGGCCTGAGTGCTACAGTTGCAGGTGTAGGATAAGTGGCTGCTGAGGGGTCACTGCTACCTGTCTGTTGCCGTTTGTAGATAAGCTCACCAGTGCCAGCAAGGACTCTCTGAGCACCCACACCAGCCCC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................TCCCACCAGAGTCgcca........................................................................................................................................................................................................ | 17 | gcca | 84.00 | 0.00 | - | 57.00 | - | 11.00 | - | 5.00 | - | 7.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GGTGTAGGATAAGTGGga......................................................................................... | 18 | ga | 37.00 | 7.00 | 21.00 | - | - | - | 11.00 | - | - | - | - | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TTCCCACCAGAGTCgcca........................................................................................................................................................................................................ | 18 | gcca | 25.00 | 0.00 | - | - | - | - | - | - | 8.00 | - | 6.00 | - | 5.00 | - | - | - | 2.00 | - | 1.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................GGTGTAGGATAAGTGGgag........................................................................................ | 19 | gag | 18.00 | 7.00 | 3.00 | - | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGGTGTAGGATAAGTGGga......................................................................................... | 19 | ga | 16.00 | 7.00 | 4.00 | - | 7.00 | 1.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................GGTGTAGGATAAGTGGgagc....................................................................................... | 20 | gagc | 10.00 | 7.00 | 1.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................GGTGTAGGATAAGTGGg.......................................................................................... | 17 | g | 8.00 | 7.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GGTGTAGGATAAGTGG........................................................................................... | 16 | 1 | 7.00 | 7.00 | 6.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGGTGTAGGATAAGTGG........................................................................................... | 17 | 1 | 7.00 | 7.00 | 3.00 | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGGTGTAGGATAAGTGGg.......................................................................................... | 18 | g | 6.00 | 7.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGGTGTAGGATAAGTG............................................................................................ | 16 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGGTGTAGGATAAGTGGgag........................................................................................ | 20 | gag | 4.00 | 7.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGGTGTAGGATAAGT............................................................................................. | 15 | 2 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GTGTAGGATAAGTGG........................................................................................... | 15 | 2 | 3.00 | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GTGTAGGATAAGTGGga......................................................................................... | 17 | ga | 2.50 | 3.00 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TTCCCACCAGAGTCTCca........................................................................................................................................................................................................ | 18 | ca | 2.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TCCCACCAGAGTCgcc......................................................................................................................................................................................................... | 16 | gcc | 2.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TAAGCTCACCAGTGCCAGCAAGGACTC...................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GGTGTAGGATAAGTG............................................................................................ | 15 | 6 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGCCAGCAAGGACTCTCTGAGCACCC........... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................AGGTGTAGGATAAGTGGgagc....................................................................................... | 21 | gagc | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TAAGCTCACCAGTGCCAGCAAGGACTCTCTGt................. | 32 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................................................................................................TGCCAGCAAGGACTCTCTGAGCA.............. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CTCACCAGTGCCAGCAAGGACTCTCTGAG................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................................................................CCAGTGCCAGCAAGGACTCTCTGAGC............... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................AGGTGTAGGATAAGTGGgagg....................................................................................... | 21 | gagg | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TTCCCACCAGAGTCTCct........................................................................................................................................................................................................ | 18 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TCCCACCAGAGTCgtca........................................................................................................................................................................................................ | 17 | gtca | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TCCCACCAGAGTCgcct........................................................................................................................................................................................................ | 17 | gcct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGGTGTAGGATAAGTGG........................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GTGTAGGATAAGTGGgag........................................................................................ | 18 | gag | 0.50 | 3.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GTGTAGGATAAGTGGgagg....................................................................................... | 19 | gagg | 0.50 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TACCACAAGCTATTTGGACTTGATGGCTTCTTTTCCCACCAGAGTCTCATCCCTGATGCTAGAGTAGGTTCTTCTTGTCCAGAGGTGAAGCCTGCTCTTCCTAGTTCCCAAGTCGCATCTGTGGCCTGAGTGCTACAGTTGCAGGTGTAGGATAAGTGGCTGCTGAGGGGTCACTGCTACCTGTCTGTTGCCGTTTGTAGATAAGCTCACCAGTGCCAGCAAGGACTCTCTGAGCACCCACACCAGCCCC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................AGGATAAGTGGCTGCTtc...................................................................................... | 18 | tc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GATGCTAGAGTAGGTTCTTCTTGTCCAGA....................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |