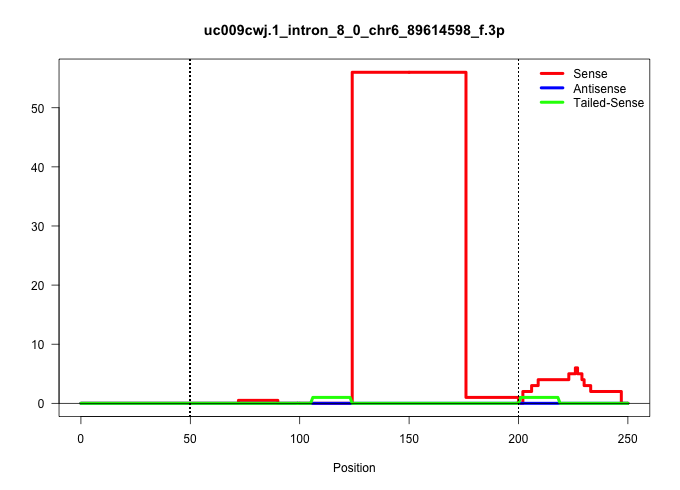
| Gene: Txnrd3 | ID: uc009cwj.1_intron_8_0_chr6_89614598_f.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(3) PIWI.mut |
(10) TESTES |
| CCGTCTCAGTGAATCCTCCATTTAATTCTATCCGCAAACTGCAGAGCCTGGATGTATTCTTTCTCTGCTTAGTCTAGAGAGCTCAAGTGTACCTAGCTACACATGTGATCATCACACTGCAAGCACACCTTTACATGCATGTCCTTTGGTCAGTTTGTTGTCTGCCATGTTAGAGCCAGTACTTCATGTGTCCTCACTAGGTTCAACAGTTGGAGAAAGGTTTACCAGGAAAATTGAAAGTCGTGGCTAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................ACACCTTTACATGCATGTCCTTTGGTCAGTTTGTTGTCTGCCATGTTAGAGC.......................................................................... | 52 | 1 | 56.00 | 56.00 | 56.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TCAACAGTTGGAGAAAGGTTTACCAGG..................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CAGTACTTCATGTGTCCTCACTAG.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................GATCATCACACTGCcagc.............................................................................................................................. | 18 | cagc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................................................................TTCAACAGTTGGAGAttt............................... | 18 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................................ACCAGGAAAATTGAAAGTCGTGGC... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TCAACAGTTGGAGAAAGGTTTACCAGGA.................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................AGGAAAATTGAAAGTCGTGGC... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................................................................CAGTTGGAGAAAGGTTTACCAGGAAAA................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................................TTGGAGAAAGGTTTACCA....................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................TCTAGAGAGCTCAAGTGT................................................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 |
| CCGTCTCAGTGAATCCTCCATTTAATTCTATCCGCAAACTGCAGAGCCTGGATGTATTCTTTCTCTGCTTAGTCTAGAGAGCTCAAGTGTACCTAGCTACACATGTGATCATCACACTGCAAGCACACCTTTACATGCATGTCCTTTGGTCAGTTTGTTGTCTGCCATGTTAGAGCCAGTACTTCATGTGTCCTCACTAGGTTCAACAGTTGGAGAAAGGTTTACCAGGAAAATTGAAAGTCGTGGCTAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|