
| Gene: Gfpt1 | ID: uc009csz.1_intron_13_0_chr6_87027235_f.5p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(14) TESTES |
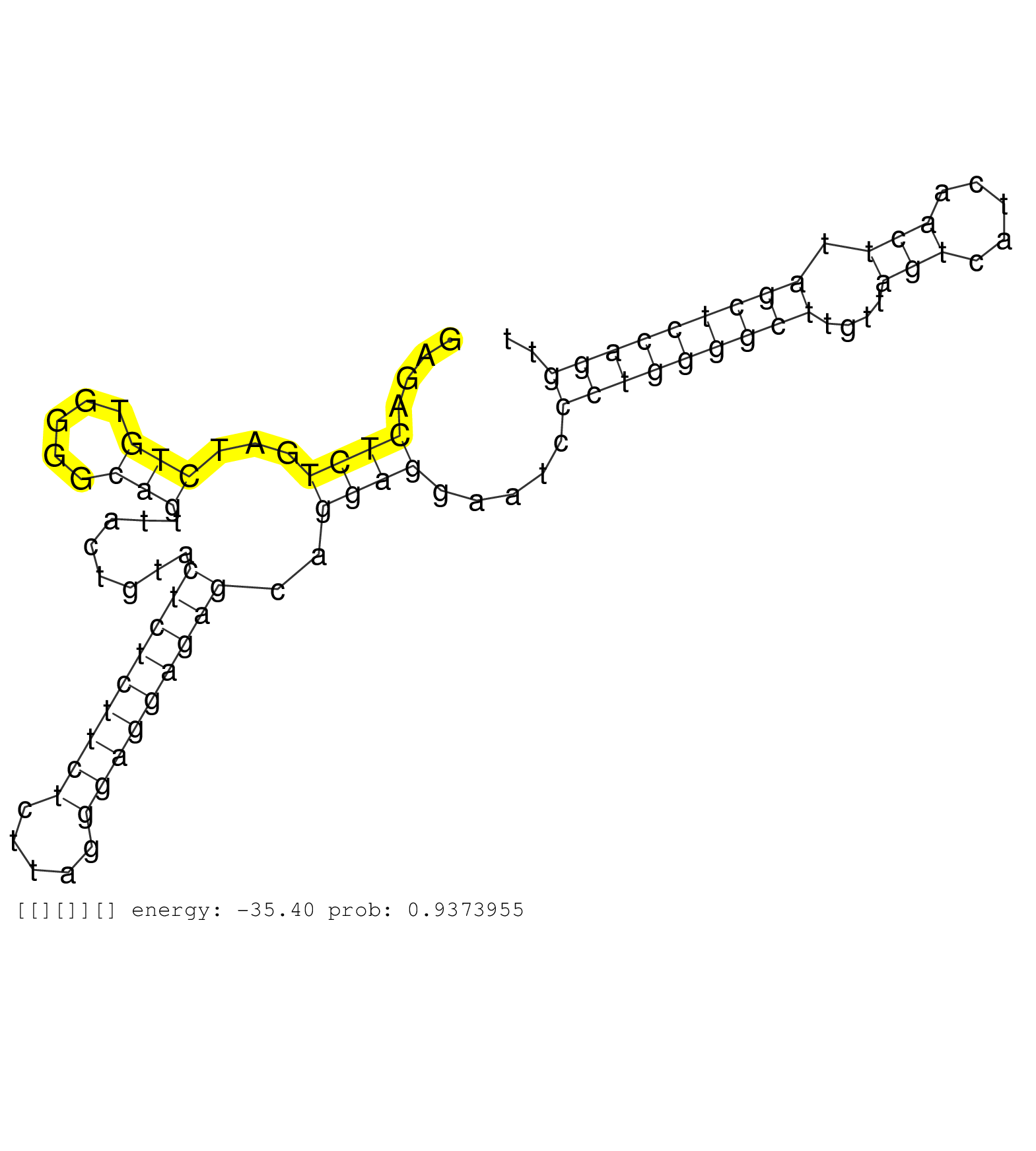
| GCGGGGTTCATATTAATGCTGGTCCTGAGATTGGCGTGGCCAGTACAAAGGTAAATTTTTGGTTTCTTCCATGGAAGTTACTTAACCATACTGTTGGTAAGAGACTCTGATCTGTGGGGCAGTTACTGTACTCTCTTCTCTTAGGGAGGAGAGCAGGAGGAATCCCTGGGGCTTGTTAGTCATCAACTTAGCTCCAGGTTCAGTGAGTGACTGTGTCTTAGGGAACACGACAGAGATTGATAGAGCAGGA ........................................................................................................((((...(((.....)))........(((((((((.....)))))))))..)))).....(((((((((....(((.....))).))))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAATTTTTGGTTTCTTCCATGGAAGTTACTTAACCATACTGTTGGTAAGA.................................................................................................................................................... | 52 | 1 | 49.00 | 49.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GCTGGTCCTGAGATTGGCGT..................................................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..........TATTAATGCTGGTCCTGAGATTGGCGTG.................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................TGAGATTGGCGTGGCCAGTACAAAGGc...................................................................................................................................................................................................... | 27 | c | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................TGAGATTGGCGTGGCCAGTACAAAGGcat.................................................................................................................................................................................................... | 29 | cat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................CTGAGATTGGCGTGGCCAGTACAAAG........................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................TGAGATTGGCGTGGCCAGTACAAAGGcata................................................................................................................................................................................................... | 30 | cata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAAATTTTTGGTTTCTTCCATGGAAGT............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........TATTAATGCTGGTCCTGAGATTGGCGTGG................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGAGATTGGCGTGGCCAGTACAAAGGcatt................................................................................................................................................................................................... | 30 | catt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................AGATTGGCGTGGCCAGTACAAAGGcata................................................................................................................................................................................................... | 28 | cata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........TATTAATGCTGGTCCTGAGATTGGCGT..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GCTGGTCCTGAGATTGGagt..................................................................................................................................................................................................................... | 20 | agt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................TCCTGAGATTGGCGTGGCCAGTACAAc......................................................................................................................................................................................................... | 27 | c | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................TGGTCCTGAGATTGGCGTGGCC................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| GCGGGGTTCATATTAATGCTGGTCCTGAGATTGGCGTGGCCAGTACAAAGGTAAATTTTTGGTTTCTTCCATGGAAGTTACTTAACCATACTGTTGGTAAGAGACTCTGATCTGTGGGGCAGTTACTGTACTCTCTTCTCTTAGGGAGGAGAGCAGGAGGAATCCCTGGGGCTTGTTAGTCATCAACTTAGCTCCAGGTTCAGTGAGTGACTGTGTCTTAGGGAACACGACAGAGATTGATAGAGCAGGA ........................................................................................................((((...(((.....)))........(((((((((.....)))))))))..)))).....(((((((((....(((.....))).))))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|