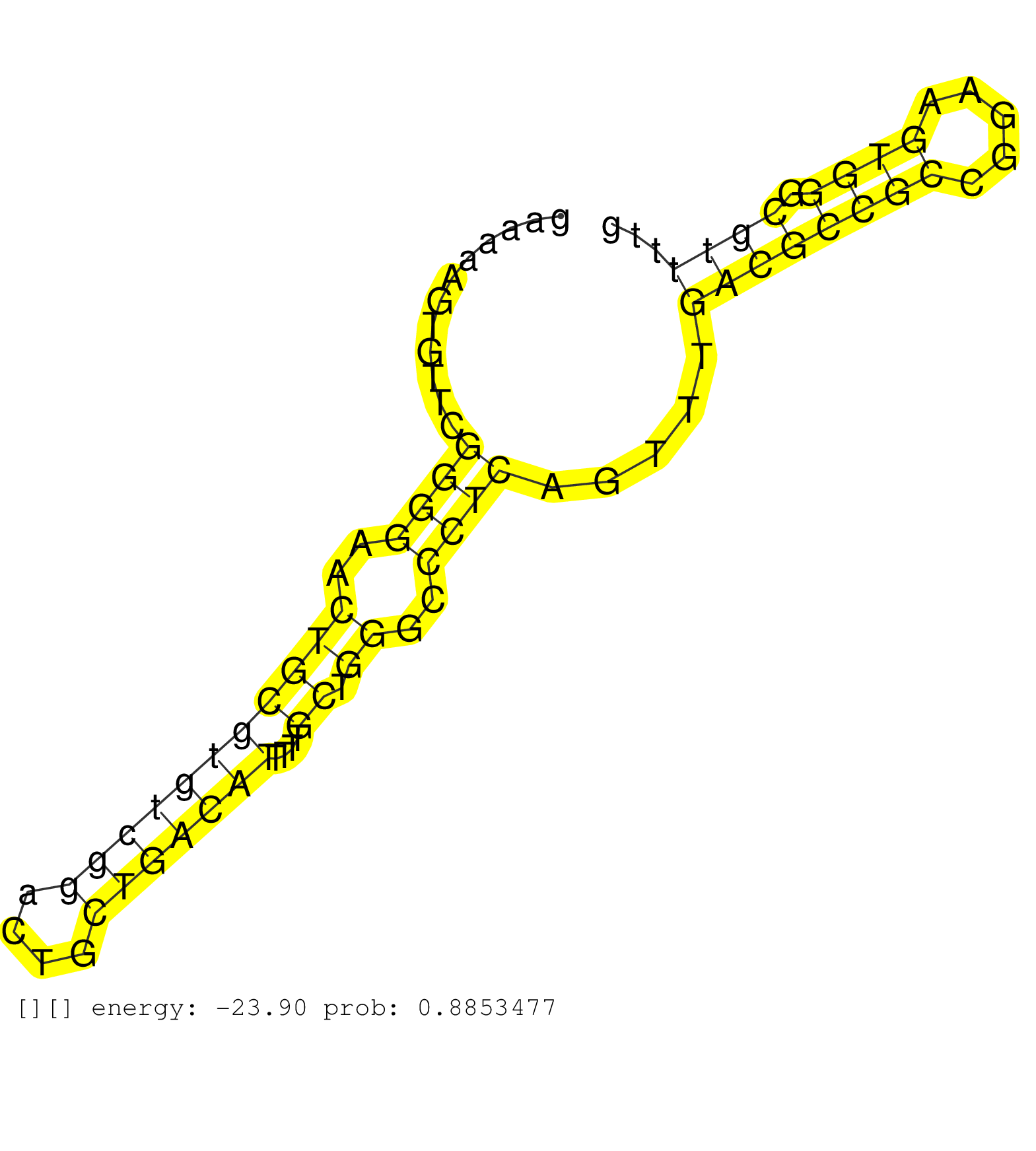

| Gene: C87436 | ID: uc009cru.1_intron_0_0_chr6_86388482_f.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| GAGACGCGGCGAGGGTCGTCACCAACGAGATTCCTAGAGTTGCGAGGCAGGTAAGGGGGCAAAGTGGGAGAGGCCAAGCACCGGCGGAGCACGGCTTAGGGTGGGAACAGAGGCGGAAAAAGTGTTCGGGGAACTGCGTGTCGGACTGCTGACATTTTTGCTGGGCCCTCAGTTTGACGCCGCCGGAAGTGGGCGTTTTGCGCAGGATGCCGGGAAGCGGGTCGCATGGGAAGTGCGTTTTCCAGCCGGC ...............................................................................................................................((((..(((((((((((....)))))))....)).))..)))).....((((((((.....)))).))))..................................................... ...................................................................................................................116.................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................CTGCTGACATTTTTGCTGGGCCCTCAGTTTGACGCCGCCGGAAGTGGGCGTT..................................................... | 52 | 1 | 169.00 | 169.00 | 169.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGTGTTCGGGGAACTGC................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TTTTGCTGGGCCCTCAGTTTGACGCC..................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GTGTTCGGGGAACTGCG................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CCTAGAGTTGCGAGGCAG........................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GTGTCGGACTGCTGACATT.............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| GAGACGCGGCGAGGGTCGTCACCAACGAGATTCCTAGAGTTGCGAGGCAGGTAAGGGGGCAAAGTGGGAGAGGCCAAGCACCGGCGGAGCACGGCTTAGGGTGGGAACAGAGGCGGAAAAAGTGTTCGGGGAACTGCGTGTCGGACTGCTGACATTTTTGCTGGGCCCTCAGTTTGACGCCGCCGGAAGTGGGCGTTTTGCGCAGGATGCCGGGAAGCGGGTCGCATGGGAAGTGCGTTTTCCAGCCGGC ...............................................................................................................................((((..(((((((((((....)))))))....)).))..)))).....((((((((.....)))).))))..................................................... ...................................................................................................................116.................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................GGGGAACTGCGTGTCGGACTGCTGACA................................................................................................ | 27 | 1 | 5.00 | 5.00 | - | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................ACCAACGAGATatag........................................................................................................................................................................................................................... | 15 | atag | 3.00 | 0.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GGTCGTCACCAACGAGATTCCTAGAGTTGC............................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................GGGGAACTGCGTGTCGGACTGCTGACATT.............................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................CGGGGAACTGCGTGTCGGACTGCTG................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GCGAGGGTCGTCACCAACGAGATTCCTA...................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................AACGAGATTCCTAGAGTTGCGAGGCA......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................GGGAACTGCGTGTCGGACTGCTGACA................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................ACGAGATTCCTAGAGTTGCGAGGCA......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............GTCGTCACCAACGAGATT.......................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................ACCAACGAGATTCCTAGAGTTGCGAGGCA......................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CGTGTCGGACTGCTGACAT............................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................GACGCCGCCGGAAcggt.............................................................. | 17 | cggt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GTCGGACTGCTGAgatc.................................................................................................. | 17 | gatc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GCCCTCAGTTTGACGCCG.................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................GGAAAAAGTGTTCGGGGAAC.................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........GCGAGGGTCGTCACCAACGAGATTCC........................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TTCGGGGAACTGCGTGTCGGACTGCTGACAtt................................................................................................ | 32 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................TCCTAGAGTTGCGAGGCAGGTA..................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GGCGAGGGTCGTCACCAACGAGATTCCTA...................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................GACATTTTTGCTGGGCCCTCAGTTTGAC........................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................GTCACCAACGAGATTCCTAGAGTTGCGAGt............................................................................................................................................................................................................ | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |