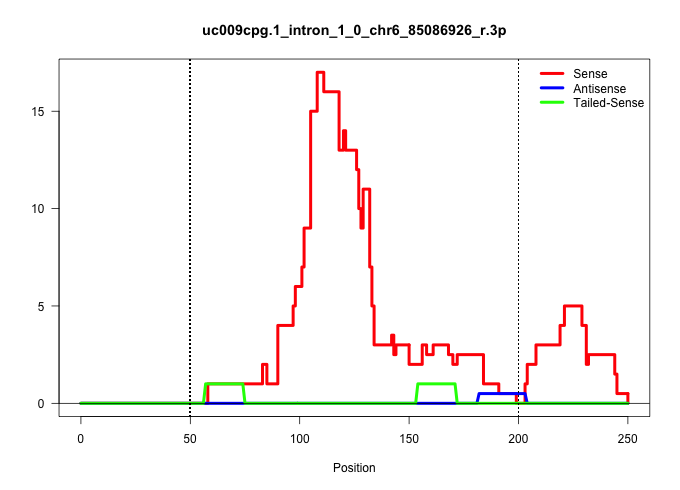
| Gene: Spr | ID: uc009cpg.1_intron_1_0_chr6_85086926_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
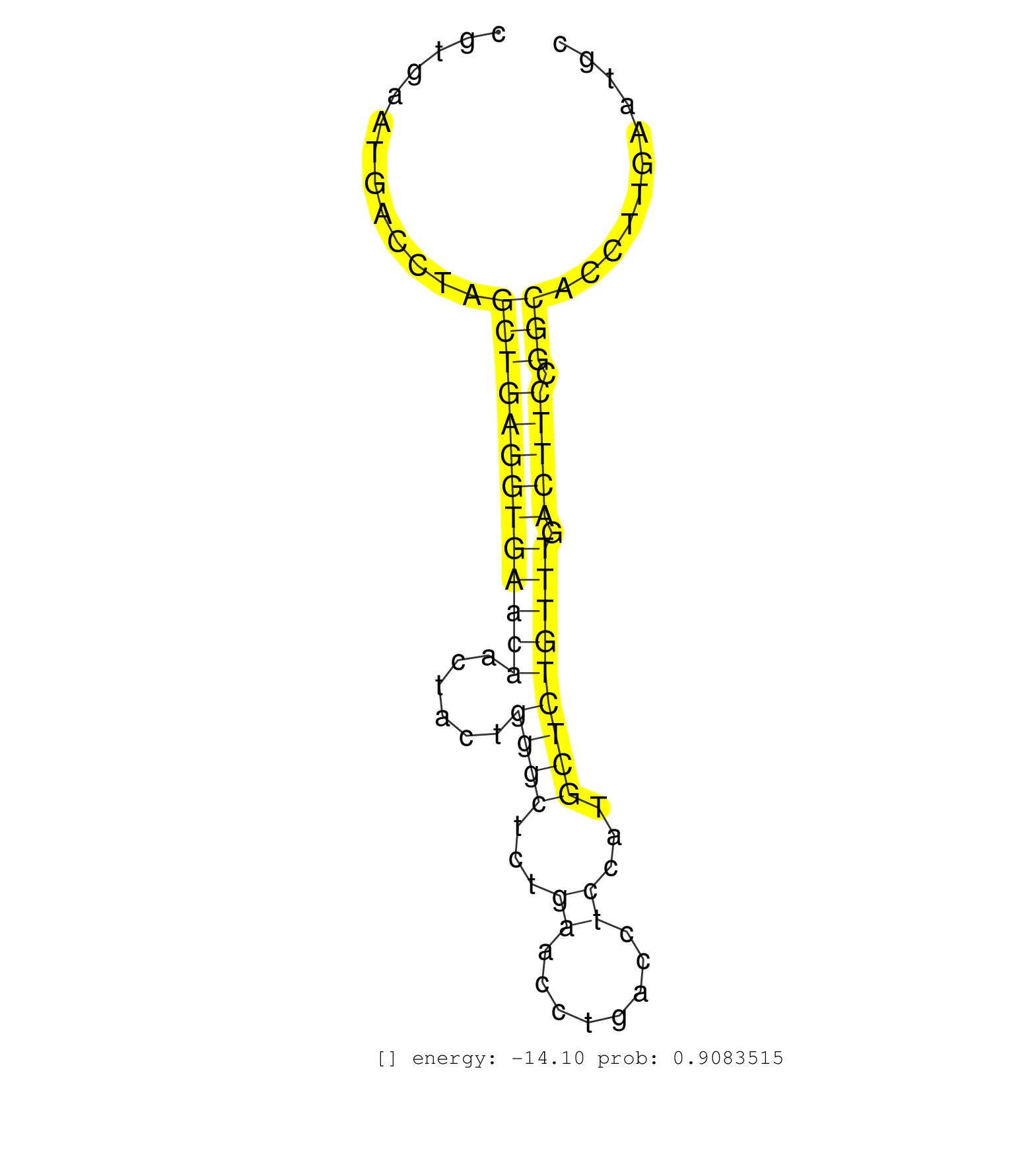
| GTTTTTTTTTTTTTTCAGCCACTCTTGGGGATGTTTCCAAAGGCTTCCTCAACGTGAATGACCTAGCTGAGGTGAACAACTACTGGGCTCTGAACCTGACCTCCATGCTCTGTTTGACTTCCGGCACCTTGAATGCCTTCCAGGATAGCCCTGGCCTGAGCAAGACTGTGGTTAACATCTCATCTCTGTGTGCCCTGCAGCCCTACAAAGGCTGGGGTCTGTACTGTGCGGGGAAGGCTGCCCGAGACAT .................................................................(((((((((((((......((((...((........))...))))))))).))))).)))............................................................................................................................. ....................................................53.................................................................................136................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................TGCTCTGTTTGACTTCCGGCACCTTGA...................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................TGAACCTGACCTCCATGCTCTGTTTGAC.................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................GAGGTGAACAACTACTGGGC.................................................................................................................................................................. | 20 | 2 | 2.50 | 2.50 | - | - | - | - | - | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AGGCTGGGGTCTGTACTGTGCGG................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TCTGTTTGACTTCCGGCACCTTGAAT.................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGCTCTGTTTGACTTCCGGCACCTTGAA..................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................ACCTCCATGCTCTGTTTGACTTCCGGCA............................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TCCATGCTCTGTTTGACTTCCGGCAC........................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CCATGCTCTGTTTGACTTCCGGCACC.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TGGGCTCTGAACCTGACCTCCATGCTCT........................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TACAAAGGCTGGGGTCTGTACTGTGC..................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................GACCTCCATGCTCTGTTTGACTTC................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................CCTGAGCAAGACTGTata.............................................................................. | 18 | ata | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CCGGCACCTTGAATGCCTTCCAG........................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGCAAGACTGTGGTTAACATCTCATC.................................................................. | 28 | 2 | 1.00 | 1.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ..........................................................TGACCTAGCTGAGGTGAACAACTACTG..................................................................................................................................................................... | 27 | 2 | 1.00 | 1.00 | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TACTGTGCGGGGAAGGCTGCCCGA..... | 24 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGAATGCCTTCCAGGATAGCCCTGGCCTG............................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGAATGCCTTCCAGGATAGCC.................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGTACTGTGCGGGGAAGGCTGCCCG...... | 25 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TCTGTTTGACTTCCGGC............................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................GGGGTCTGTACTGTGCGGGG................. | 20 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................ATGACCTAGCTGAGGatt............................................................................................................................................................................... | 18 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................CCATGCTCTGTTTGACTTCCGGCAC........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................ACAAAGGCTGGGGTCTGTACTGTGCGG................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................AAGACTGTGGTTAACATCTCATCTCTGTGT........................................................... | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ............................................................................................................................................................................TAACATCTCATCTCTGTGTGCCCTGCA................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AGCAAGACTGTGGTTAACATCTCATC.................................................................. | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................GAAGGCTGCCCGAGACAT | 18 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATAGCCCTGGCCTGAGCAAGACTGTG................................................................................ | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ..............................................................................................................................................GGATAGCCCTGGCCTGAGCAAGACTG.................................................................................. | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| GTTTTTTTTTTTTTTCAGCCACTCTTGGGGATGTTTCCAAAGGCTTCCTCAACGTGAATGACCTAGCTGAGGTGAACAACTACTGGGCTCTGAACCTGACCTCCATGCTCTGTTTGACTTCCGGCACCTTGAATGCCTTCCAGGATAGCCCTGGCCTGAGCAAGACTGTGGTTAACATCTCATCTCTGTGTGCCCTGCAGCCCTACAAAGGCTGGGGTCTGTACTGTGCGGGGAAGGCTGCCCGAGACAT .................................................................(((((((((((((......((((...((........))...))))))))).))))).)))............................................................................................................................. ....................................................53.................................................................................136................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................TCTCTGTGTGCCCTGCAGCCCT.............................................. | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |