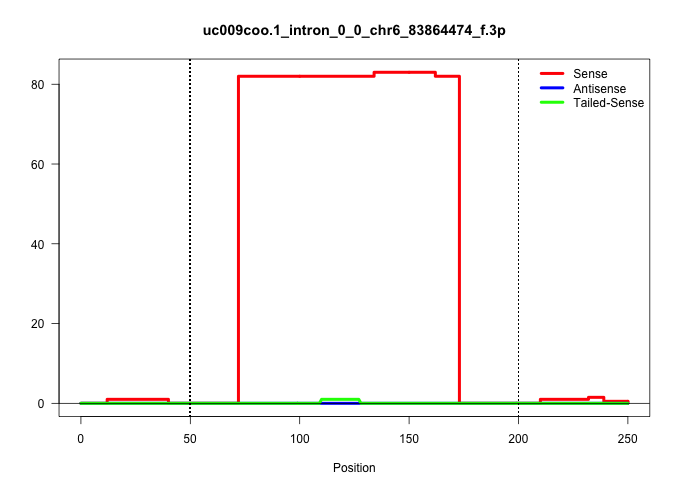
| Gene: Znf638 | ID: uc009coo.1_intron_0_0_chr6_83864474_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| TGGGGTTTGTGTTCTGGAATCAGAAAAGAAGAGAAAGTTTTAGAGGAAATTTTAAATCTTTGTAAGCCCTTACTCTAAAAGTATATGCAATGCAAAATGATATCATGGCTTGATTTCAAAATGTTTTACTCTGGCTGTAGAACCCACTGCATGAAGCAGTTTGAGTTAAAATGATCTAATATTTGTGTTTTAACTTTCAGCTTTGTGTTATTCTTGGAAAATTTCGCACCACTTGTGAATTCCTTGAGCC .....................................................................................................((((.....))))........(((((((.......)))))))..(((((.....)))))..((((((((((..............))))))))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................CTCTAAAAGTATATGCAATGCAAAATGATATCATGGCTTGATTTCAAAATGT.............................................................................................................................. | 52 | 1 | 105.00 | 105.00 | 105.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGTAGAACCCACTGCATGA................................................................................................ | 19 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TACTCTGGCTGTAGAACCCACTGCATGAAGCA............................................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............TCTGGAATCAGAAAAGAAGAGAAAGTTT.................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................TGATTTCAAAATGTctgg.......................................................................................................................... | 18 | ctgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TTCTTGGAAAATTTCGCACCACTTGTGAA........... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................CTGTAGAACCCACTGCATGAAGCAGTTT........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................TAGAACCCACTGCATGAAGC............................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................TGATTTCAAAATGTTTTACTCTGGC................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................................................................................................................TTGTGAATTCCTTGAGCC | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| TGGGGTTTGTGTTCTGGAATCAGAAAAGAAGAGAAAGTTTTAGAGGAAATTTTAAATCTTTGTAAGCCCTTACTCTAAAAGTATATGCAATGCAAAATGATATCATGGCTTGATTTCAAAATGTTTTACTCTGGCTGTAGAACCCACTGCATGAAGCAGTTTGAGTTAAAATGATCTAATATTTGTGTTTTAACTTTCAGCTTTGTGTTATTCTTGGAAAATTTCGCACCACTTGTGAATTCCTTGAGCC .....................................................................................................((((.....))))........(((((((.......)))))))..(((((.....)))))..((((((((((..............))))))))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................TCTGGCTGTAGAACcgtc........................................................................................................... | 18 | cgtc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................AAGTATATGCAATGCAActt........................................................................................................................................................... | 20 | ctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................AGCCCTTACTCTAAAAgttc.......................................................................................................................................................................... | 20 | gttc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |