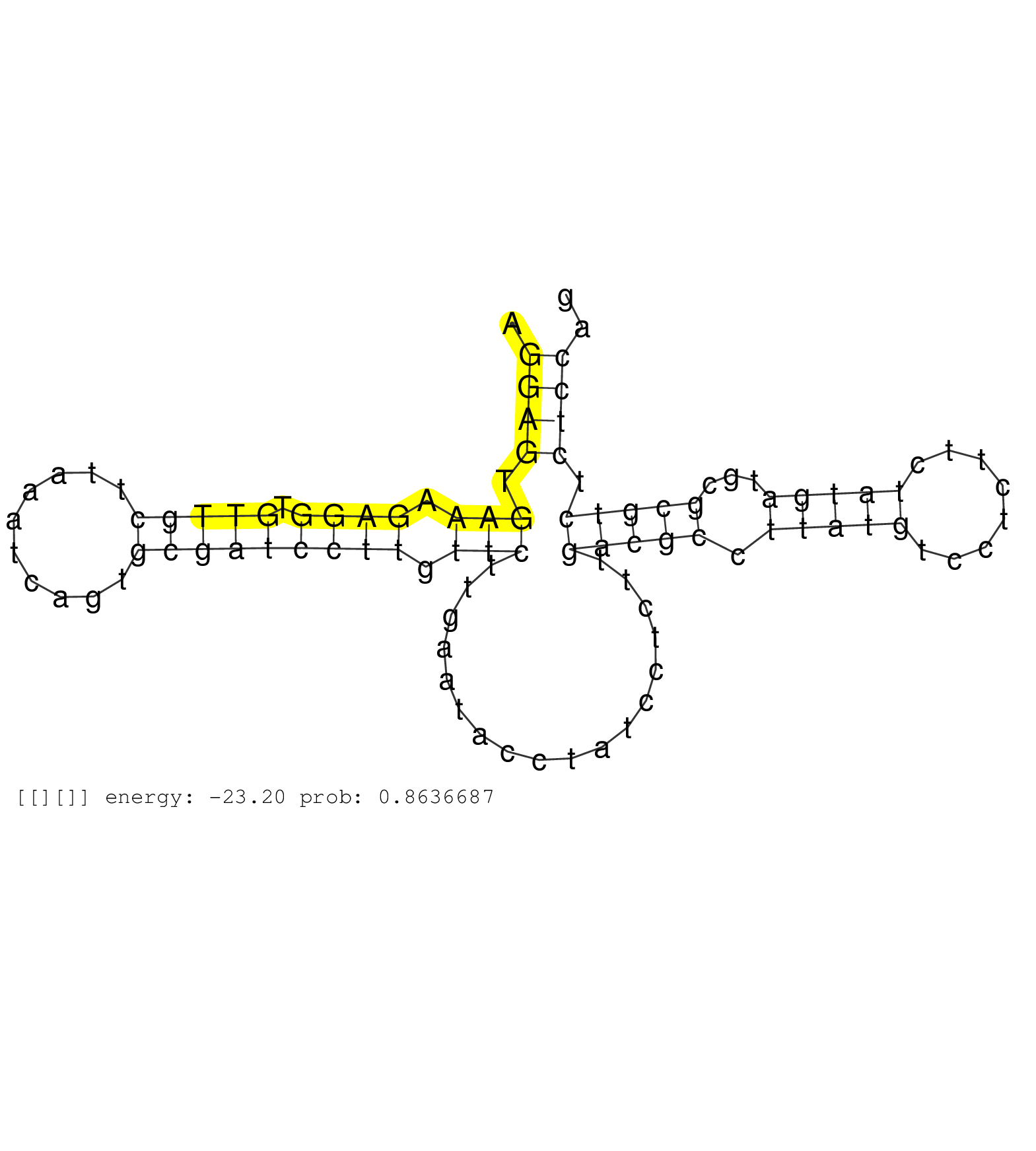

| Gene: Wdr54 | ID: uc009cmv.1_intron_7_0_chr6_83105535_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(12) TESTES |
| AGCTCCACGTTAAAGAGGGTGCTGGAGTGAGTCCCCCACTTATCACTCAGGTAAGGCAAGGAGTGAAAGAGGTGTTGCTTAAATCAGTGCGATCCTTGTTCTTGAATACCTATCCTCTTGACGCCTTATGTCCTCTTCTATGATGCGCGTCTCTCCAGGTTCACTGGTGTGTCCTCCCCTTCAGAGTACTGCTGGTCCTCACCTCGCA ...........................................................((((.(((.((((.(((((..........))))))))).)))..................(((((.(((((........)))))...))))).)))).................................................... ..........................................................59.................................................................................................158................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGGCAAGGAGTGAAAGAGGTGTT.................................................................................................................................... | 26 | 1 | 12.00 | 12.00 | 4.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 |
| ..................................................GTAAGGCAAGGAGTGAAAGAGGTGT..................................................................................................................................... | 25 | 1 | 9.00 | 9.00 | 5.00 | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - |
| ..................................................GTAAGGCAAGGAGTGAAAGAGGTGTTG................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAAGGCAAGGAGTGAAAGAGGTGc..................................................................................................................................... | 25 | c | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGCAAGGAGTGAAAGAGGTG...................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGCAAGGAGTGAAAGAGGcgt..................................................................................................................................... | 25 | cgt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGCAAGGAGTGAAAGAGGTGTa.................................................................................................................................... | 26 | a | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGCAAGGAGTGAAAGAGGggtg.................................................................................................................................... | 26 | ggtg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| AGCTCCACGTTAAAGAGGGTGCTGGAGTGAGTCCCCCACTTATCACTCAGGTAAGGCAAGGAGTGAAAGAGGTGTTGCTTAAATCAGTGCGATCCTTGTTCTTGAATACCTATCCTCTTGACGCCTTATGTCCTCTTCTATGATGCGCGTCTCTCCAGGTTCACTGGTGTGTCCTCCCCTTCAGAGTACTGCTGGTCCTCACCTCGCA ...........................................................((((.(((.((((.(((((..........))))))))).)))..................(((((.(((((........)))))...))))).)))).................................................... ..........................................................59.................................................................................................158................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|