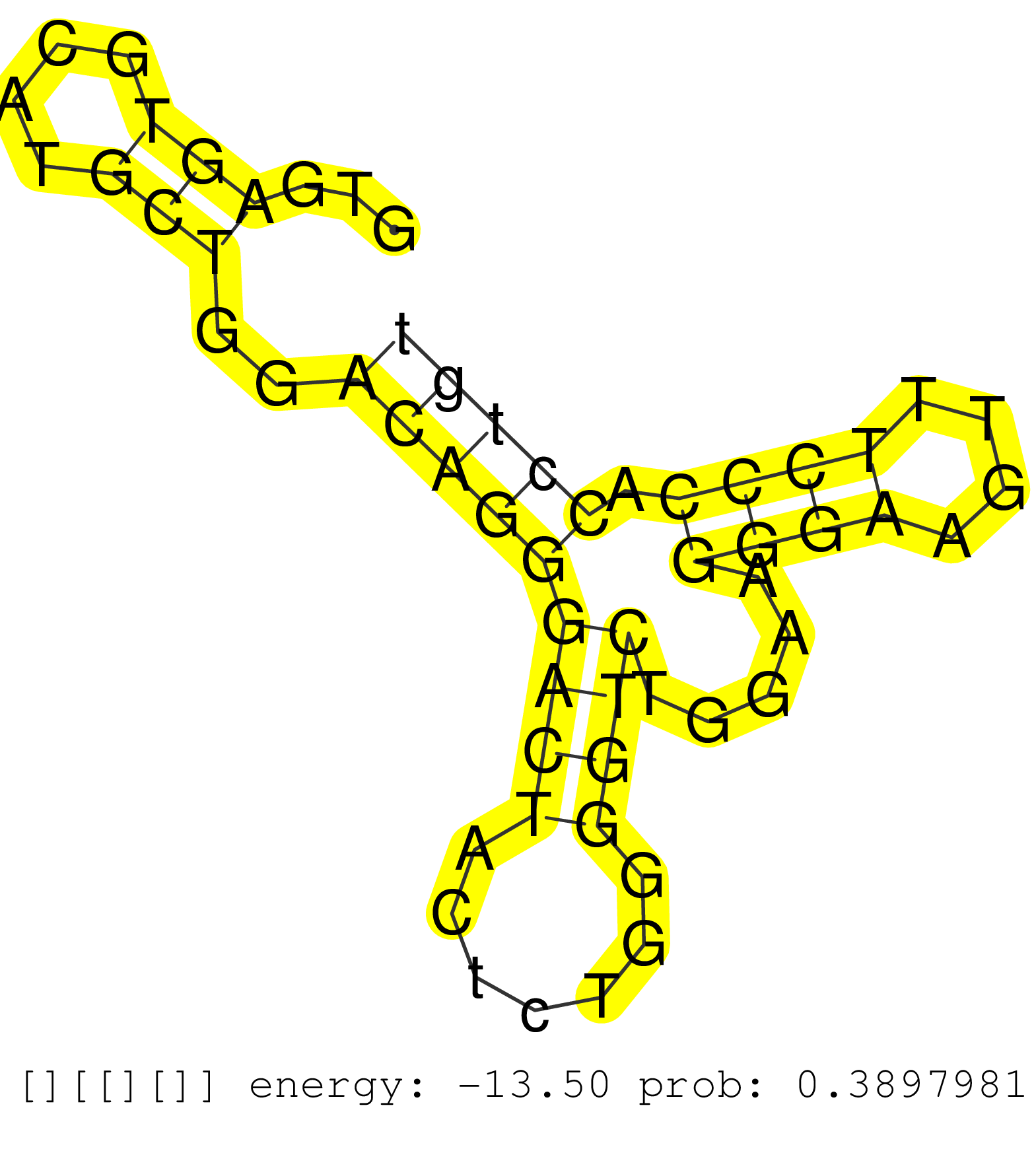

| Gene: Aup1 | ID: uc009cly.1_intron_4_0_chr6_83005965_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| ACGTGGTACAACCTCTTACCCTGCAAGTTCAGAGACCCCTGGTCTCTGTGGTGAGTGCATGCTGGACAGGGACTACTCTGGGGTCTGGAAGGGAAGTTTCCCACCTGTGGCCCTGGCTTAGACCTCACTTCATGCCCTCCCCTCAGACGGTGTCAGATGCCTCCTGGGTCTCAGAACTGCTGTGGTCCCTTTTTGT .....................................................(((....)))..(((((((((.......)))).....((((....)))).)))))........................................................................................ ..................................................51.......................................................108...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTGCATGCTGGACAGGGACTAC........................................................................................................................ | 26 | 1 | 32.00 | 32.00 | 11.00 | 5.00 | 6.00 | 2.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - |
| ..................................................GTGAGTGCATGCTGGACAGGGACTA......................................................................................................................... | 25 | 1 | 8.00 | 8.00 | 4.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGCATGCTGGACAGGGACTAC........................................................................................................................ | 25 | 1 | 8.00 | 8.00 | - | 1.00 | 1.00 | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGTGCATGCTGGACAGGGACT.......................................................................................................................... | 24 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ...................................................TGAGTGCATGCTGGACAGGGACTACTC...................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - |
| ..................................................GTGAGTGCATGCTGGACAGGGACTACT....................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGCATGCTGGACAGGGACTACTCTGG................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGCATGCTGGACAGGGACT.......................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................TGGGGTCTGGAAGGGAAGTTTCCCAC............................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGCATGCTGGACAGGGACTACTC...................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGCATGCTGGACAGGGAatac........................................................................................................................ | 26 | atac | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGCATGCTGGACAGGGccta......................................................................................................................... | 25 | ccta | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGCATGCTGGACAGGGACTACTCTGG................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................CCTCCTGGGTCTCAGAACTG................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................GGACTACTCTGGGGTCTGGAAGGG....................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................CGGTGTCAGATGCCTCCTGG............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGCATGCTGGACAGGGA............................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................TGGTCTCTGTGGTGAGTGCATGCTG.................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TGGAAGGGAAGTTTCCCACCTGTGGC..................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ACGTGGTACAACCTCTTACCCTGCAAGTTCAGAGACCCCTGGTCTCTGTGGTGAGTGCATGCTGGACAGGGACTACTCTGGGGTCTGGAAGGGAAGTTTCCCACCTGTGGCCCTGGCTTAGACCTCACTTCATGCCCTCCCCTCAGACGGTGTCAGATGCCTCCTGGGTCTCAGAACTGCTGTGGTCCCTTTTTGT .....................................................(((....)))..(((((((((.......)))).....((((....)))).)))))........................................................................................ ..................................................51.......................................................108...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................GACGGTGTCAGATGCCTCCTGGGTCTCA....................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |