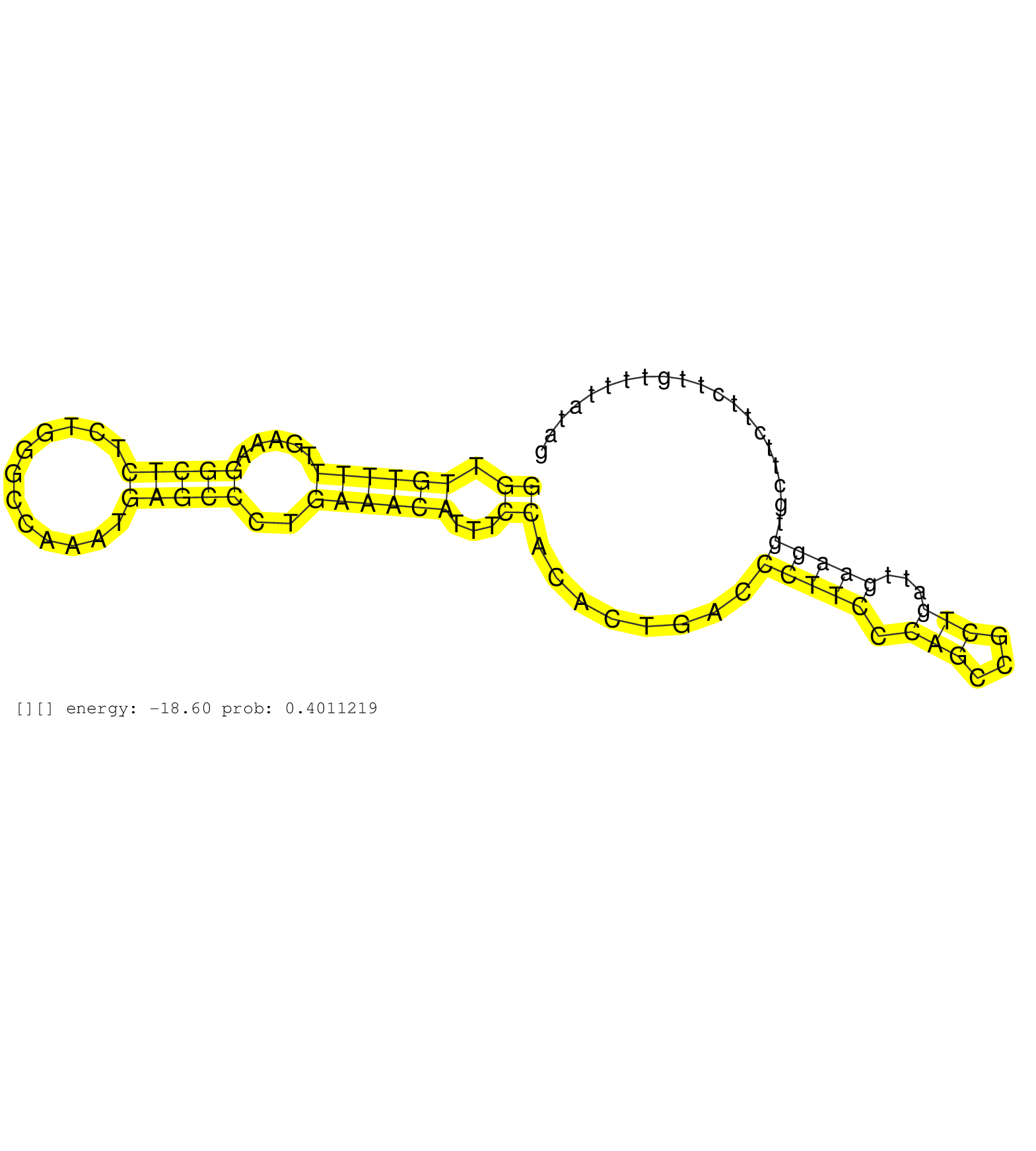
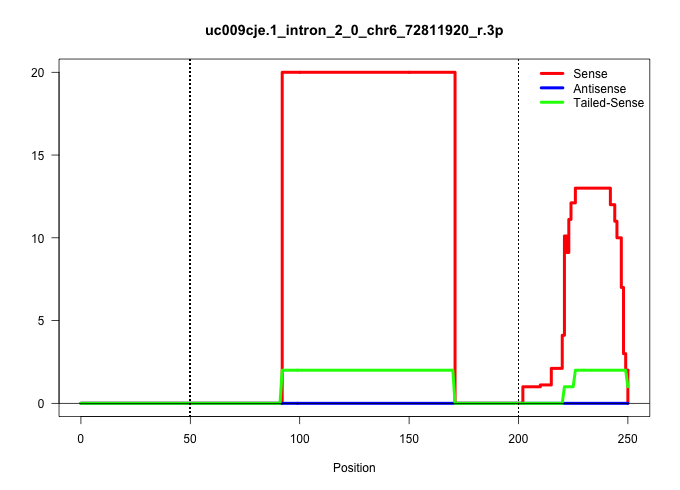
| Gene: Kcmf1 | ID: uc009cje.1_intron_2_0_chr6_72811920_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| CCGCCACCCGTCTTGAGGTCCTCTTCATTAGGATTGACCTTTCTGAGTAAGGAGTTTTCCACACTGCTCAGATTACTGTGTTTTTTACTGTGTTGTTGCTGGTTGTTTTTGAAAGGCTCTCTGGGCCAAATGAGCCCTGAAACATTTCCACACTGACCCTTCCCAGCCGCTGATTGAAGGTGCTTCTTCTTGTTTTATAGGTGTCAGCTGTGATGCATGTTTAAAAGGAAATTTTCGAGGTCGCAGATAT ....................................................................................................((.((((((.....(((((............)))))..))))))...))........(((((.(((...)))...)))))...................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................TTGTTGCTGGTTGTTTTTGAAAGGCTCTCTGGGCCAAATGAGCCCTGAAACA.......................................................................................................... | 52 | 1 | 23.00 | 23.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TAAAAGGAAATTTTCGAGGTCGCAGAT.. | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................AAAGGAAATTTTCGAGGTCGCAGATAT | 27 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TAAAAGGAAATTTTCGAGGTCGCAGA... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................................TAAAAGGAAATTTTCGAGGTCGCAGATA. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................GGAAATTTTCGAGGTC........ | 16 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................CATGTTTAAAAGGAAATTTTCGAGGTCGC...... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................GGAAATTTTCGAGGTCGCAGATATt | 25 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TAAAAGGAAATTTTCGAGGTCGCAGctat | 29 | ctat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................................................GTCAGCTGTGATGCATGTTT............................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................................................AAGGAAATTTTCGAGGTCGCAGAT.. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................................................................................TTAAAAGGAAATTTTCGAGGTCGCA..... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TTAAAAGGAAATTTTCGAGGTCGCAGA... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TGATGCATGTTTAAAA........................ | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| CCGCCACCCGTCTTGAGGTCCTCTTCATTAGGATTGACCTTTCTGAGTAAGGAGTTTTCCACACTGCTCAGATTACTGTGTTTTTTACTGTGTTGTTGCTGGTTGTTTTTGAAAGGCTCTCTGGGCCAAATGAGCCCTGAAACATTTCCACACTGACCCTTCCCAGCCGCTGATTGAAGGTGCTTCTTCTTGTTTTATAGGTGTCAGCTGTGATGCATGTTTAAAAGGAAATTTTCGAGGTCGCAGATAT ....................................................................................................((.((((((.....(((((............)))))..))))))...))........(((((.(((...)))...)))))...................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................AAATGAGCCCTGAAActat............................................................................................................ | 19 | ctat | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTTCTTCTTGTTTTAggaa..................................................... | 19 | ggaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |