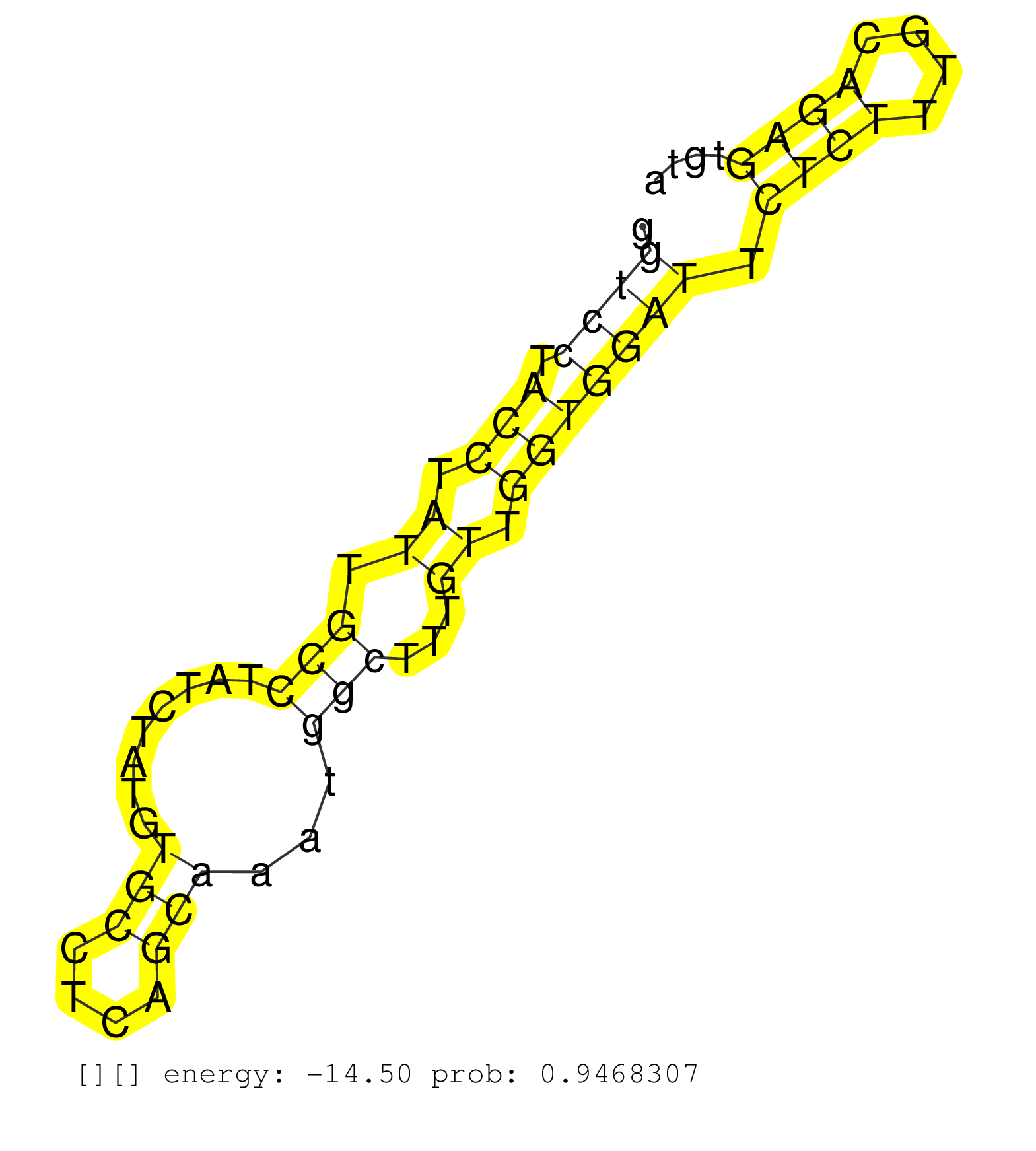
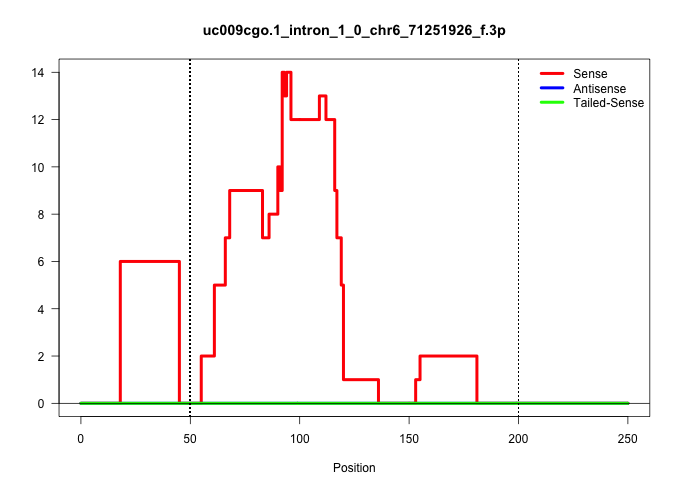
| Gene: Gm1070 | ID: uc009cgo.1_intron_1_0_chr6_71251926_f.3p | SPECIES: mm9 |
 |
 |
|
(6) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| CTTTCCCTGGTGTGCCCTTAGTTACAAAAACAGGGAGTTGCTAGGGAAAAGGTCCTACCTATTGCCTATCTATGTGCCTCAGCAAATGGCTTTGTTGGTGGATTCTCTTTGCAGAGTGTAAATCCTGAGTGGTTGGGGAAAGGACCCTCAGGCTCCCCCGCTCTGAGTTATGCCTCCCCAGCCCCACCCATGGAGCCCAGCCCCAGGCAGGTCAACTGCCACCTGTAGGTCACAGCCAGCCACCTGCTCG ...................................................((((.(((.((.(((........(((....)))...)))...)).))))))).((((....))))...................................................................................................................................... ..................................................51...................................................................120................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................TAGTTACAAAAACAGGGAGTTGCTAGG............................................................................................................................................................................................................. | 27 | 1 | 6.00 | 6.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - |
| ..........................................................................................TTTGTTGGTGGATTCTCTTTGCAGAG...................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGTTGGTGGATTCTCTTTGCAGAGTGTA.................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................TGTTGGTGGATTCTCTTTGCAGAGTGT................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ....................................................................TCTATGTGCCTCAGCAAATGGCTTTGTT.......................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TACCTATTGCCTATCTATGTGCCTCAGC....................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................TTGCCTATCTATGTGCCTCAGCAAATGGCT............................................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CCCCGCTCTGAGTTATGCCTCCCCAG..................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTGGTGGATTCTCTTTGCAGAGTGTA.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TATCTATGTGCCTCAGCAAATGGCTTT............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CTTTGTTGGTGGATTCTCTTTGCAGAGT..................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................TATCTATGTGCCTCAGCAAATGG................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TGGCTTTGTTGGTGGATTCTCTTTGC.......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................TTGCCTATCTATGTGCCTCAGCAAATGGC................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TTGTTGGTGGATTCTCTTTGCAGAGT..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TCCCCCGCTCTGAGTTATGCCTCCCCAG..................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................TGCAGAGTGTAAATCCTGAGTGGTTGG.................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| CTTTCCCTGGTGTGCCCTTAGTTACAAAAACAGGGAGTTGCTAGGGAAAAGGTCCTACCTATTGCCTATCTATGTGCCTCAGCAAATGGCTTTGTTGGTGGATTCTCTTTGCAGAGTGTAAATCCTGAGTGGTTGGGGAAAGGACCCTCAGGCTCCCCCGCTCTGAGTTATGCCTCCCCAGCCCCACCCATGGAGCCCAGCCCCAGGCAGGTCAACTGCCACCTGTAGGTCACAGCCAGCCACCTGCTCG ...................................................((((.(((.((.(((........(((....)))...)))...)).))))))).((((....))))...................................................................................................................................... ..................................................51...................................................................120................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................CCCGCTCTGAGTTATGtga.............................................................................. | 19 | tga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |