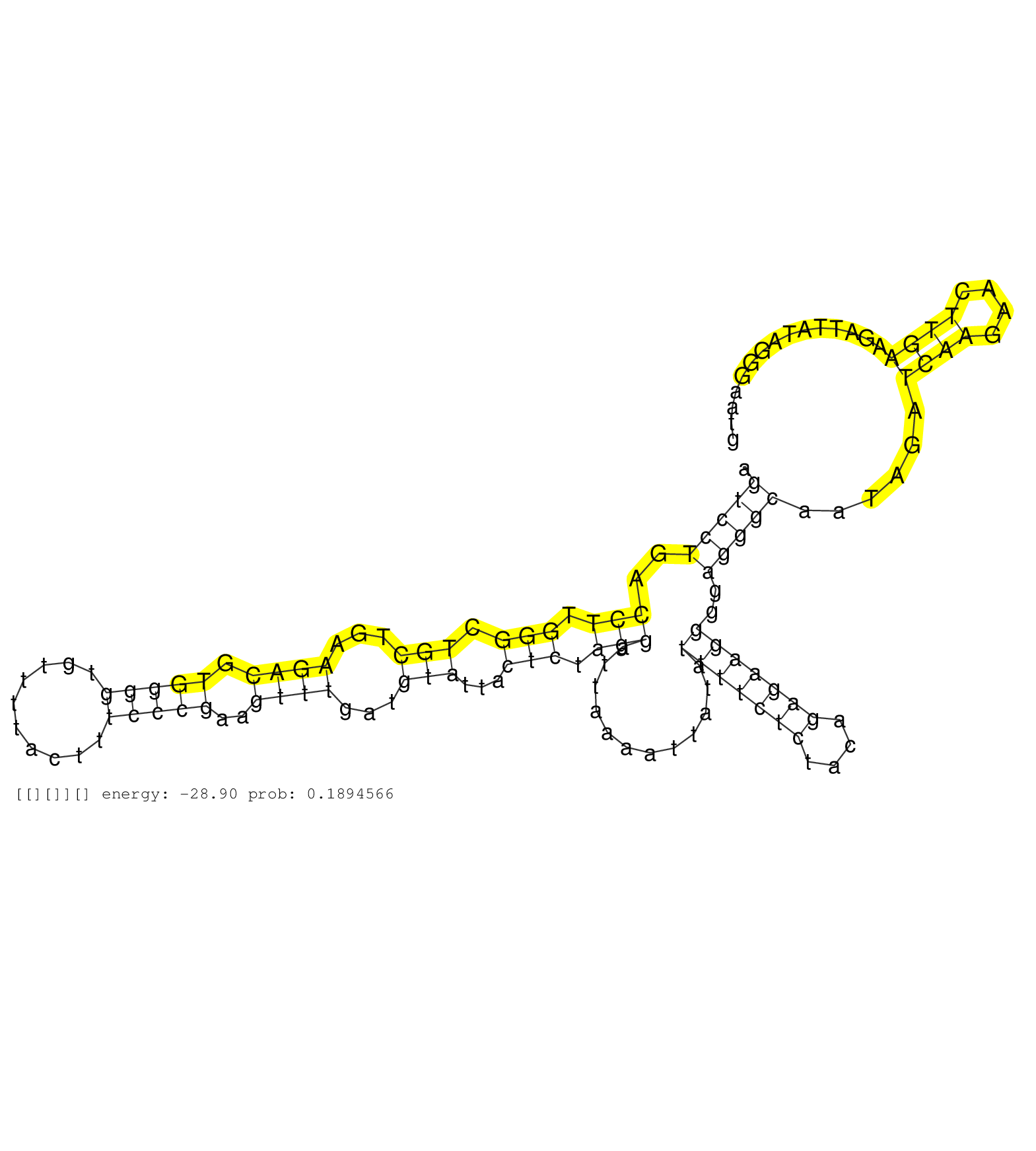

| Gene: Herc3 | ID: uc009cde.1_intron_21_0_chr6_58844619_f.5p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| AAGAAGTTGATACCCGGGGGAGACAGAGTGGCCGTGTGCAAAGACAACAGGTTAGTCCTGACCTTGGGCTGCTGAAGACGTGGGGTGTTTTACTTTCCCGAAGTTTGATGTATTACTCTAGGATTAAAATTATATTTTCTCTACAGAGAAGGGGAGGGCAATAGATCAAGAACTTGAAGATTATAGGGAATGTTTAATAGCATAGAATTTAGTGGATAGCTCATTTATCTACAGGTCTGCATCCTTTTGG ......................................................(((((..(((.(((.(((...((((.(((((..........)))))..))))...)))...))).)))............(((((((....)))))))..)))))......((((....))))......................................................................... .....................................................54........................................................................................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................TAGATCAAGAACTTGAAGATTATAGGG.............................................................. | 27 | 1 | 10.00 | 10.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 |
| ..........................................................TGACCTTGGGCTGCTGAAGACGTG........................................................................................................................................................................ | 24 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - |
| .................................................................................................................................................................TAGATCAAGAACTTGAAGATTATAGG............................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................TGGGCTGCTGAAGACGTGGGGT.................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAGATCAAGAACTTGAAGATTATAGGGA............................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CAGAGAAGGGGAGGGCAATAGATCAA................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................CAAGAACTTGAAGATTATAGGGAATGTaa....................................................... | 29 | aa | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................TGACCTTGGGCTGCTGAAGACGTGG....................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................TCAAGAACTTGAAGATTATAGGGAA............................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................TCCCGAAGTTTGATGTATTACTCTA.................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................ATAGGGAATGTTTAATAGCATAGAATT......................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TAGGGAATGTTTAATAGCATAGAATTT........................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................CAAGAACTTGAAGATTATAGGGAATGTT........................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| AAGAAGTTGATACCCGGGGGAGACAGAGTGGCCGTGTGCAAAGACAACAGGTTAGTCCTGACCTTGGGCTGCTGAAGACGTGGGGTGTTTTACTTTCCCGAAGTTTGATGTATTACTCTAGGATTAAAATTATATTTTCTCTACAGAGAAGGGGAGGGCAATAGATCAAGAACTTGAAGATTATAGGGAATGTTTAATAGCATAGAATTTAGTGGATAGCTCATTTATCTACAGGTCTGCATCCTTTTGG ......................................................(((((..(((.(((.(((...((((.(((((..........)))))..))))...)))...))).)))............(((((((....)))))))..)))))......((((....))))......................................................................... .....................................................54........................................................................................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|