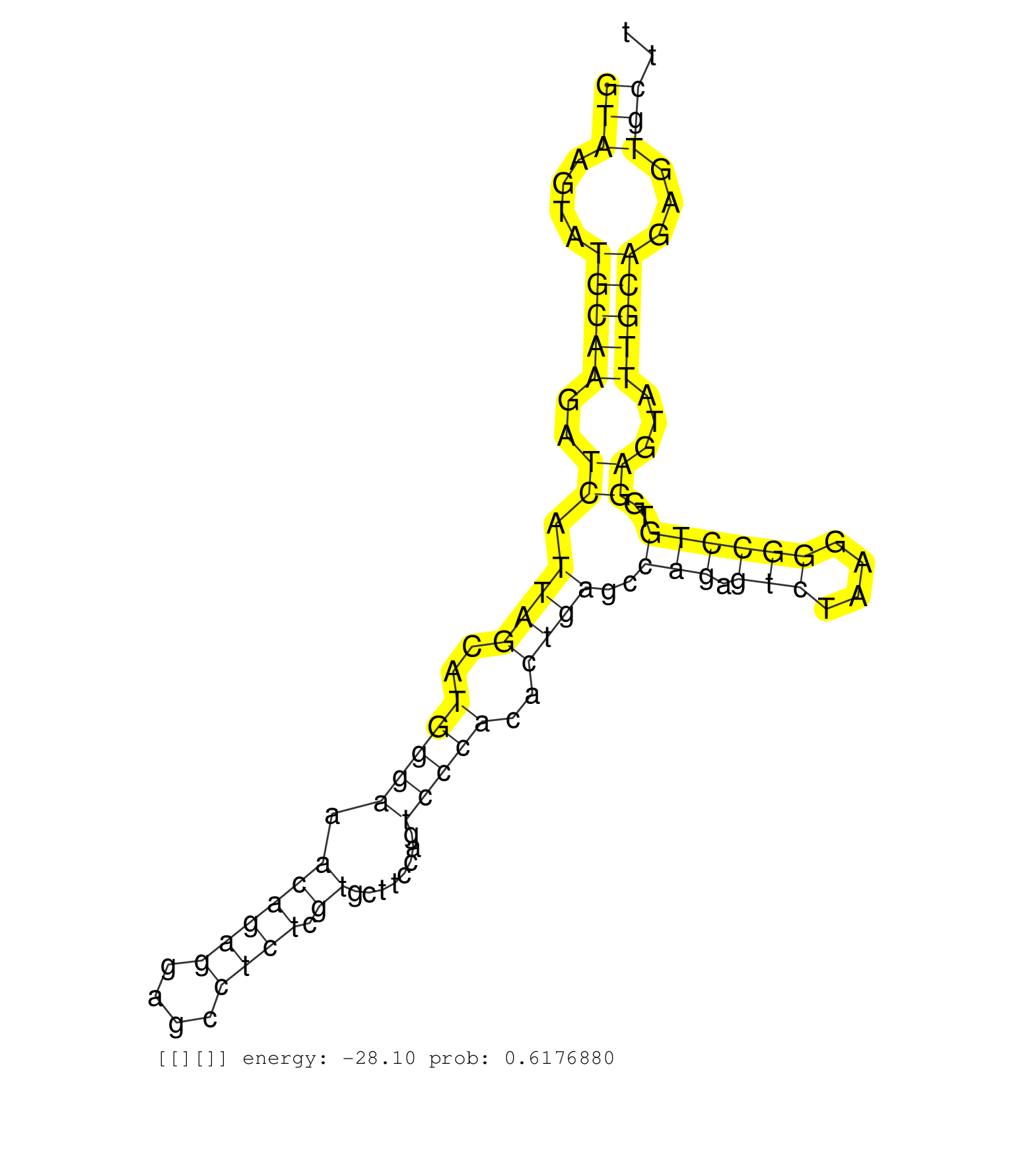
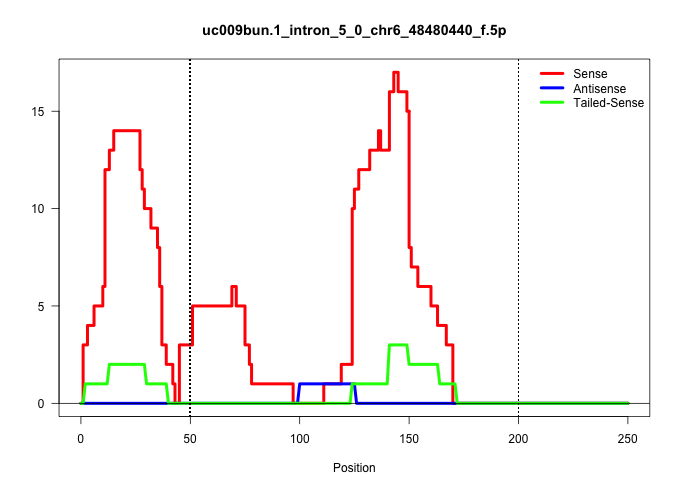
| Gene: BC068148 | ID: uc009bun.1_intron_5_0_chr6_48480440_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(14) TESTES |
| CTTCAGCCACATCTATGCCTGTGCCCGGGAGCCTACCGGCTTCCATGCCAGTAAGTATGCAAGATCATTAGCATGGGAAACAGAGGAGCCTCTCGTGCTTCCAGTCCCACACTGAGCCAGAGTCTAAGGGCCTGTGGAGTATTGCAGAGTGCTTGAGCCAGAGACTCAGCCCCTACTTCCTGTCCATTTGTGGTTTCAGGGCCGTCAGGGTTCCCGCAGAGCCTCAGTCACCCTGATACTCAAGCTCACT ..................................................(((....(((((..((.((((..(((((.((((((....)))).))........)))))..))))..(((.(((....))))))..))...)))))...))).................................................................................................. ..................................................51.....................................................................................................154.............................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................TAAGGGCCTGTGGAGTATTGCAGAGT.................................................................................................... | 26 | 1 | 6.00 | 6.00 | - | - | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTGCAGAGTGCTTGAGCCAGAGACTCAGC................................................................................ | 29 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 |
| ...........TCTATGCCTGTGCCCGGGAGCCTACC..................................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .TTCAGCCACATCTATGCCTGTGCCCG............................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGCCAGTAAGTATGCAAGATCATTAGCATG............................................................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................AGCATGGGAAACAGAGGAGCCTCTCGTG......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTGCAGAGTGCTTGAGCCAGAGACTC................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GCAGAGTGCTTGAGCCAGAGACTCAGC................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........TCTATGCCTGTGCCCGGGAGCCTACCGG................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................TTGCAGAGTGCTTGAGCCAGAGACTCAcccc.............................................................................. | 31 | cccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................GGGCCTGTGGAGTATTGCAGAGTGCTT................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............TATGCCTGTGCCCGGGAGCCTACCGGCTT................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...CAGCCACATCTATGCCTGTGCCCGGG............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TTCAGCCACATCTATGCCTGTGCCCGG.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TCTATGCCTGTGCCCGGGAGCCTA....................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TAAGGGCCTGTGGAGTATTGCAGAG..................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AAGGGCCTGTGGAGTATTGCAGAGT.................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........TCTATGCCTGTGCCCGGGAGCCTAC...................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GAGTATTGCAGAGTGCTTGAGCCAGAG....................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......CCACATCTATGCCTGTGCCCGGGAGC.......................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTATGCAAGATCATTAGCATGGGA............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..TCAGCCACATCTATGCCTGTGCCCGGGt............................................................................................................................................................................................................................ | 28 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TATGCCTGTGCCCGGGAGCCTACCGGt.................................................................................................................................................................................................................. | 27 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTATGCAAGATCATTAGCATGGG............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TAAGGGCCTGTGGAGTATTGCAGAGTG................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................CTGAGCCAGAGTCTAAGGGCCTGTGG................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGTGGAGTATTGCAGAGTGCTTGAGCCA.......................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTGCAGAGTGCTTGAGCCAGAGt...................................................................................... | 23 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGCCAGTAAGTATGCAAGATCATTAG................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........ATCTATGCCTGTGCCCGGGAGCCTAC...................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TAAGGGCCTGTGGAGTATTGCAGAGa.................................................................................................... | 26 | a | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................GAGTCTAAGGGCCTGTGGAGTATTGC......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGCCTGTGCCCGGGAGCCTACCGGCTTC............................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTTCAGCCACATCTATGCCTGTGCCCGGGAGCCTACCGGCTTCCATGCCAGTAAGTATGCAAGATCATTAGCATGGGAAACAGAGGAGCCTCTCGTGCTTCCAGTCCCACACTGAGCCAGAGTCTAAGGGCCTGTGGAGTATTGCAGAGTGCTTGAGCCAGAGACTCAGCCCCTACTTCCTGTCCATTTGTGGTTTCAGGGCCGTCAGGGTTCCCGCAGAGCCTCAGTCACCCTGATACTCAAGCTCACT ..................................................(((....(((((..((.((((..(((((.((((((....)))).))........)))))..))))..(((.(((....))))))..))...)))))...))).................................................................................................. ..................................................51.....................................................................................................154.............................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....CATCTATGCCTGTGcaga................................................................................................................................................................................................................................... | 18 | caga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................CCAGTCCCACACTGAGCCAGAGTCTA............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GTGCTTCCAGTCCCACACT......................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |