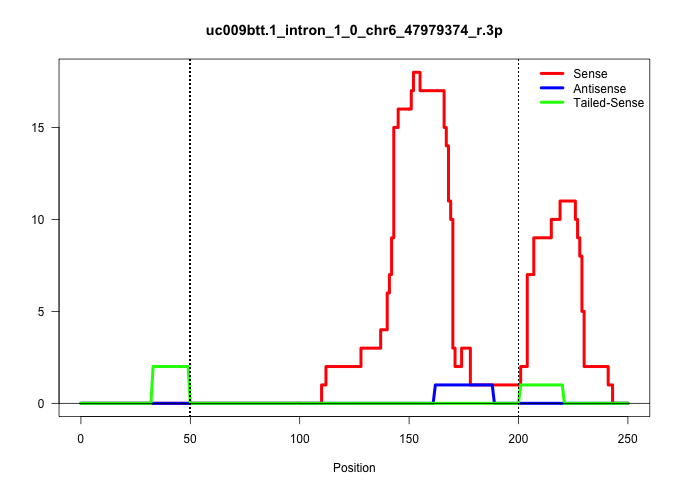
| Gene: Zfp777 | ID: uc009btt.1_intron_1_0_chr6_47979374_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(20) TESTES |
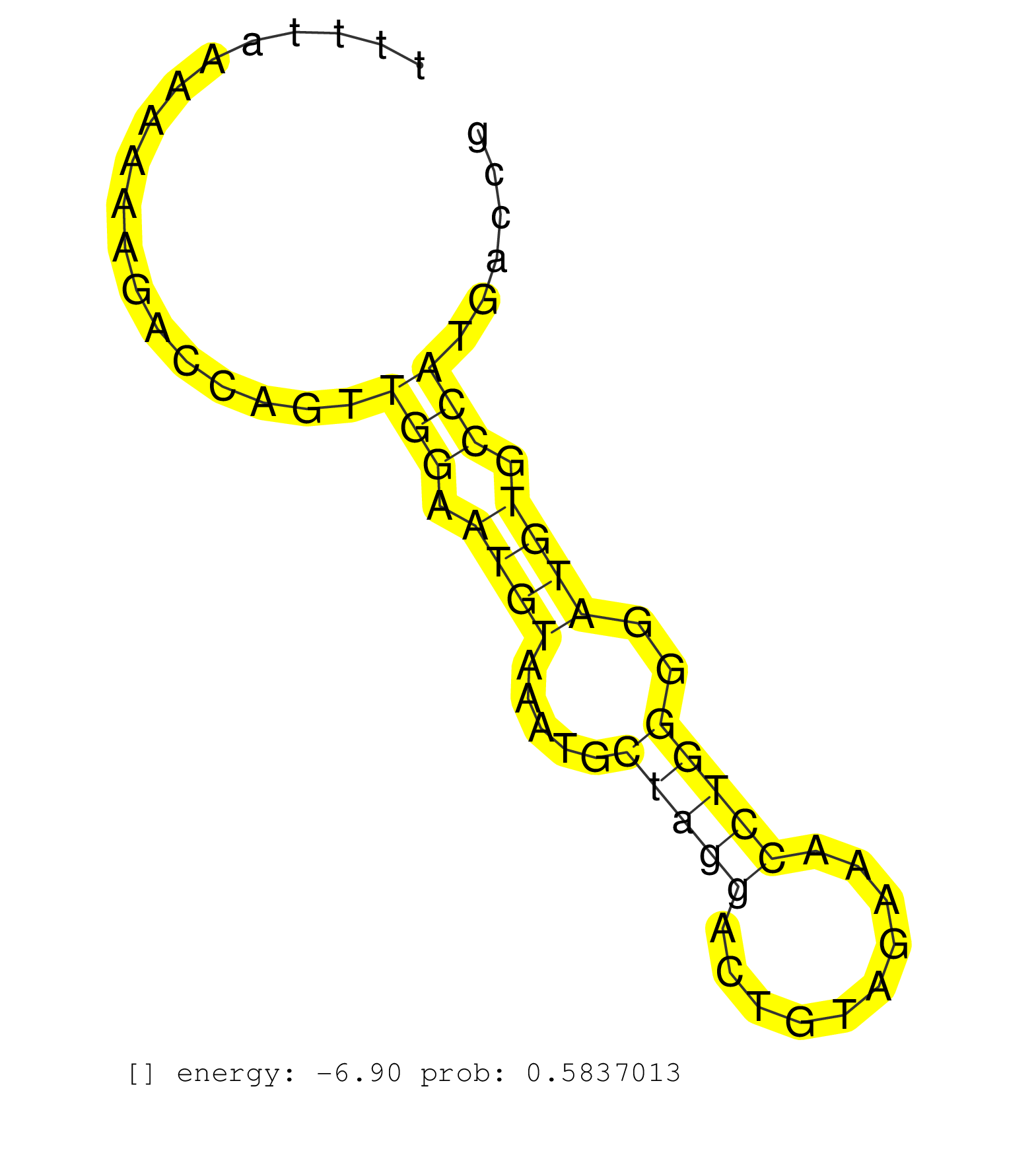
| ATCTATTGTAACTTGACTCCTGTTTGTATTAGCATTCAAGACTACTACGAAGTGTCCAGAAAATTCTTTCTGCTCACCATTAACTCTATTGTATTTTTTAGGGTCTCTTTTAAAAAAAGACCAGTTGGAATGTAAATGCTAGGACTGTAGAAACCTGGGGATGTGCCATGACCGCTCTGTGTGTCCTCACCTTCCCTCAGCACATGATGGCATCGTGATTAAGATCGAGGTGCAGACCAACGATGAGGGC .............................................................................................................................(((.((((.....(((((..........)))))..)))).))).................................................................................. ...........................................................................................................108...............................................................174.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................ACTGTAGAAACCTGGGGATGTGCCATG................................................................................ | 27 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | 3.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGATGGCATCGTGATTAAGATCGAG..................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................................TGGCATCGTGATTAAGATCGAGGTGC................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGATGGCATCGTGATTAAGATCGAGG.................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGGCATCGTGATTAAGATC........................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGTAGAAACCTGGGGATGTGCCATGA............................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................AAAAAAGACCAGTTGGAATGTAAATGC............................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAGGACTGTAGAAACCTGGGGATGTGC.................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAAAAAAAGACCAGTTGGAATGTAAATGC............................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................ACATGATGGCATCGTGATTAAGATCGAGG.................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAGGACTGTAGAAACCTGGGGATGTGCCA.................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................ACATGATGGCATCGTGATTAAGATCGA...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TAAGATCGAGGTGCAGACCAACGA....... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AATGTAAATGCTAGGACTGTAGAAACC............................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................GACTGTAGAAACCTGGGGATGTGCCATG................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................GACTGTAGAAACCTGGGGATGTGCCAT................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGACTGTAGAAACCTGGGGATGTGCC................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................ACCTGGGGATGTGCCATGACCGCTCT........................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................GGACTGTAGAAACCTGGGGATGTGCCA.................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCTAGGACTGTAGAAACCTGGGGATGTGC.................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................ATTCAAGACTACTAtga........................................................................................................................................................................................................ | 17 | tga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGATTAAGATCGAGGTGCAGACCAAC......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................ACATGATGGCATCGTGgcct............................. | 20 | gcct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................................................................................AGGACTGTAGAAACCTGGGGATGTGCCA.................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................CTCTGTGTGTCCTCACCTTCCCTCAGC................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................AACCTGGGGATGTGCCATGACCGCTCT........................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGGCATCGTGATTAAGATCG....................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................ATTCAAGACTACTctga........................................................................................................................................................................................................ | 17 | ctga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ATCTATTGTAACTTGACTCCTGTTTGTATTAGCATTCAAGACTACTACGAAGTGTCCAGAAAATTCTTTCTGCTCACCATTAACTCTATTGTATTTTTTAGGGTCTCTTTTAAAAAAAGACCAGTTGGAATGTAAATGCTAGGACTGTAGAAACCTGGGGATGTGCCATGACCGCTCTGTGTGTCCTCACCTTCCCTCAGCACATGATGGCATCGTGATTAAGATCGAGGTGCAGACCAACGATGAGGGC .............................................................................................................................(((.((((.....(((((..........)))))..)))).))).................................................................................. ...........................................................................................................108...............................................................174.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................GGACTGTAGAAAgacg................................................................................................. | 16 | gacg | 65.00 | 0.00 | 35.00 | 10.00 | 10.00 | - | - | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TAGGACTGTAGAAcgat.................................................................................................. | 17 | cgat | 6.00 | 0.00 | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CTAGGACTGTAGAAatc.................................................................................................. | 17 | atc | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................GTGCCATGACCGCTCTGTGTGTCCTCA............................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GGACTGTAGAAACgaca................................................................................................ | 17 | gaca | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CTAGGACTGTAGAAaaga.................................................................................................. | 18 | aaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |