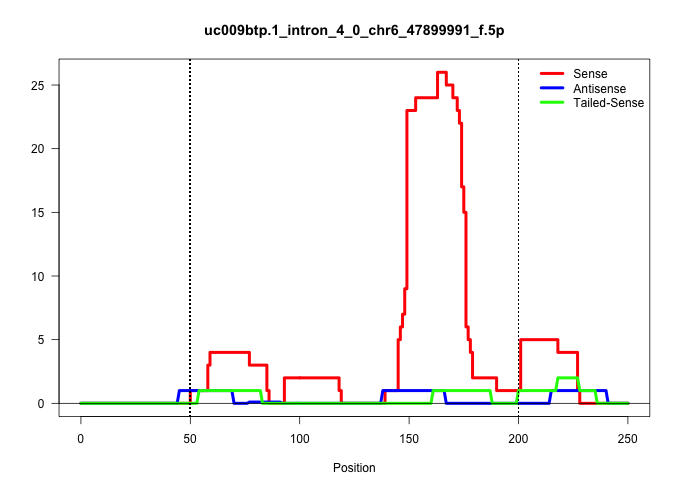
| Gene: mKIAA4190 | ID: uc009btp.1_intron_4_0_chr6_47899991_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| AGAGTTGCCACCCTCCCCTTCCACAGATAGCCTGCTTTCTGTGGAGCCAGGTGAGTGAAAAGAGAAAACACTAGTGACTGTGCCAGAGCCTCATCGAAGCTCAGAGGGCCTTTGGTAGTAACAGTCAGCCCGTGGTCAACTGGCCTGAATGGAAAATTACTGCTTGAAGACATGCTGCAGATAGAAGACTCTATCTGCTAGTGATAGAAAGGCTGTTCATATATGATCTGCTCCAGTCCTACGGTAACCC .....................................................((((...........))))..((((((((((((((.(((.((........))))).)))))))....)))))))..((((((((....))))...)))).......((((.............))))...................................................................... .....................................................54............................................................................................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................TGGAAAATTACTGCTTGAAGACATGCT.......................................................................... | 27 | 1 | 9.00 | 9.00 | 6.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................CTGCTTTCTGTGGAGCCAGGTGA.................................................................................................................................................................................................... | 23 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGATAGAAAGGCTGTTCATATATGAT....................... | 26 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGGAAAATTACTGCTTGAAGACATGC........................................................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGAGAAAACACTAGTGACTGTGCCA..................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGAATGGAAAATTACTGCTTGAAGACATG............................................................................ | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGCTTTCTGTGGAGCCAG........................................................................................................................................................................................................ | 18 | 2 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................ATGGAAAATTACTGCTTGAAGACATG............................................................................ | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TATCTGCTAGTGATAGAAAGGCTGTTC................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GTGATAGAAAGGCTGTTCATATATGATt...................... | 28 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGAATGGAAAATTACTGCTTGAAGACA.............................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TCGAAGCTCAGAGGGCCTTTGGTAG.................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................GCTTGAAGACATGCTGCAGATAGAAGt.............................................................. | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................AAATTACTGCTTGAAGACATGCTGCA....................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................GAATGGAAAATTACTGCTTGAAGA................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................ATATATGATCTGCTaaga.............. | 18 | aaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................CTGGCCTGAATGGAAAATTACTGCTTGA................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................TCGAAGCTCAGAGGGCCTTTGGTAGT................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TTGAAGACATGCTGCAGATAGAAGACT............................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TTGAAGACATGCTGCAGATAGAAGACTC........................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGAAAAGAGAAAACACTAGTGA............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGGAAAATTACTGCTTGAAGACATGCTGCA....................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGATAGAAAGGCTGTTCATATATGATC...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGAATGGAAAATTACTGCTTGAAGACAT............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGAAAAGAGAAAACACTAGTGACTGggg....................................................................................................................................................................... | 29 | ggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AAGAGAAAACACTAGTGACTGTGCCAG.................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................AATGGAAAATTACTGCTTGAAGACATG............................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGGAAAATTACTGCTTGAAGACATGCTG......................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGGAAAATTACTGCTTGAAGACATGCTGC........................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| AGAGTTGCCACCCTCCCCTTCCACAGATAGCCTGCTTTCTGTGGAGCCAGGTGAGTGAAAAGAGAAAACACTAGTGACTGTGCCAGAGCCTCATCGAAGCTCAGAGGGCCTTTGGTAGTAACAGTCAGCCCGTGGTCAACTGGCCTGAATGGAAAATTACTGCTTGAAGACATGCTGCAGATAGAAGACTCTATCTGCTAGTGATAGAAAGGCTGTTCATATATGATCTGCTCCAGTCCTACGGTAACCC .....................................................((((...........))))..((((((((((((((.(((.((........))))).)))))))....)))))))..((((((((....))))...)))).......((((.............))))...................................................................... .....................................................54............................................................................................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................TATATGATCTGCTCCAGTCCTA......... | 22 | 1 | 5.00 | 5.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ACTGGCCTGAATGGAAAATTACTGCTTGA................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................GCCAGGTGAGTGAAAAGAGAAAACA.................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GTTCATATATGATCTGCTCCAGTCCTA......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................................................TTCATATATGATCTGCTCCAGTCCTA......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................CTGTGCCAGAGCCTC.............................................................................................................................................................. | 15 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |