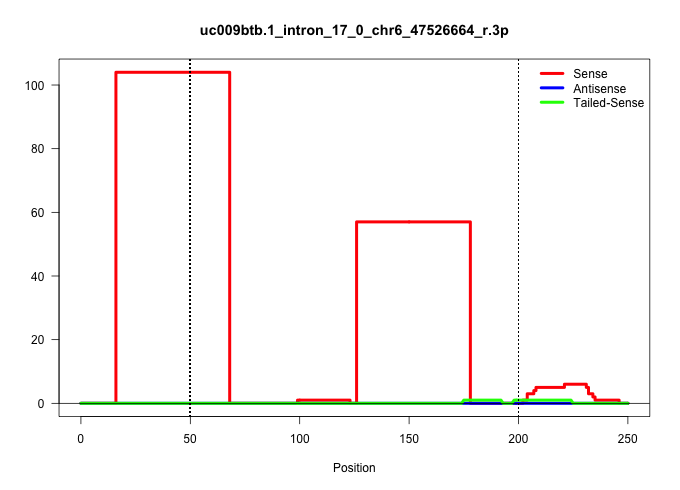
| Gene: Ezh2 | ID: uc009btb.1_intron_17_0_chr6_47526664_r.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(8) TESTES |
| TTGTTTCCCTTTATTTACAAGCAAGTAGGAGTAATGTTTTTGTATTTATTATTAGAGTGTGGAGAATTGTAGTATCTTGTGTGAATCTGTGTACATTTGAGGACATGGTCATGGTACTTCTGGTGAGTCACTAGATTTTAACCGGCTTTAAAGATGCTACATATTGTAACAATGAACAATTTCTCCTTTGTTTTTAAAAGACTATGTTTAGTTCCAATCGTCAGAAAATTTTGGAAAGAACTGAAACCTT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................ACAAGCAAGTAGGAGTAATGTTTTTGTATTTATTATTAGAGTGTGGAGAATT...................................................................................................................................................................................... | 52 | 1 | 104.00 | 104.00 | 104.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................GTCACTAGATTTTAACCGGCTTTAAAGATGCTACATATTGTAACAATGAACA........................................................................ | 52 | 1 | 57.00 | 57.00 | 57.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGTTTAGTTCCAATCGTCAGAAAATTTT.................. | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - |
| ......................................................................................................................................................................................................AGACTATGTTTAGTTCCAATCGTCAGt......................... | 27 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CAGAAAATTTTGGAAAGAACTGAAA.... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................................TTAGTTCCAATCGTCAGAAAATTTTGGA............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................AGGACATGGTCATGGTACTTCTGG............................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................................................................................TATGTTTAGTTCCAATCGTCAGAAAATTT................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TAGTTCCAATCGTCAGAAAATTTTGG................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................ACAATTTCTCCTTTcact......................................................... | 18 | cact | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - |
| TTGTTTCCCTTTATTTACAAGCAAGTAGGAGTAATGTTTTTGTATTTATTATTAGAGTGTGGAGAATTGTAGTATCTTGTGTGAATCTGTGTACATTTGAGGACATGGTCATGGTACTTCTGGTGAGTCACTAGATTTTAACCGGCTTTAAAGATGCTACATATTGTAACAATGAACAATTTCTCCTTTGTTTTTAAAAGACTATGTTTAGTTCCAATCGTCAGAAAATTTTGGAAAGAACTGAAACCTT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|