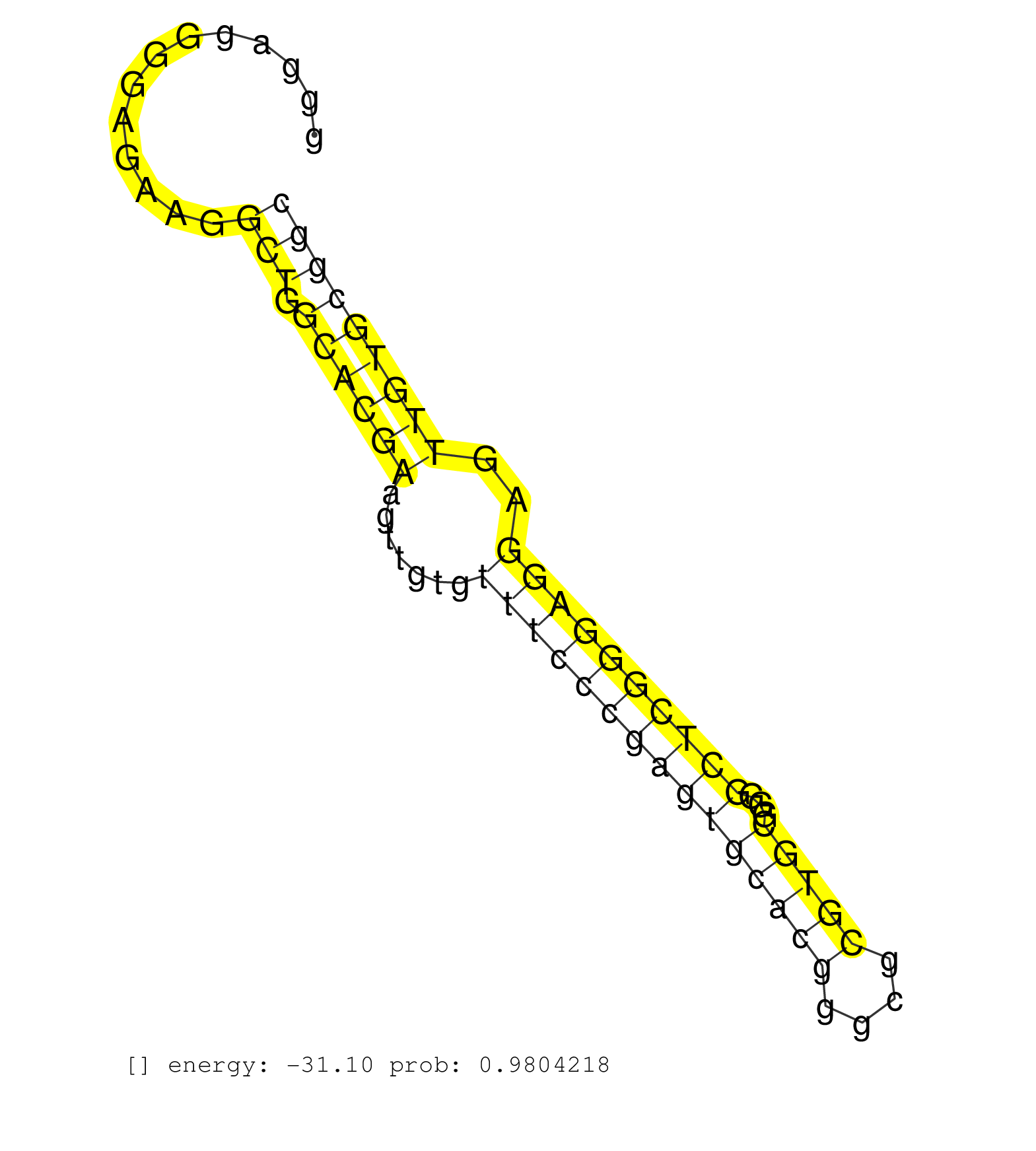

| Gene: Ssbp1 | ID: uc009bmr.1_intron_0_0_chr6_40421459_f.5p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| GGCTCCGTGCGCCGGAAGCGAGCCGCTGGCGGTCGGGCTCTGCGTGTCCGGTGAGGGAGGGGAGAAGGCTGGCACGAAGTTGTGTTTCCCGAGTGCACGGGCGCGTGCGGGGCTCGGGAGGAGTTGTGCGGCTCCTCGCTGGAGTCCGAGTGTCCCCCGCCGTTCCCTGTGCTGCTCGCGAGCGCCACTTTGGGGGAAGAGCAGCCTCTTGCCCCCGCGGCGAGGCTGGGGAGCCGGGCTTACTCAGCGC ...................................................................(((.((((((.......(((((((((((((((....)))))...))))))))))..)))))))))...................................................................................................................... ......................................................55...........................................................................132.................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................CGGTCGGGCTCTGCG.............................................................................................................................................................................................................. | 15 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CGTGCGGGGCTCGGGAGGAGTTGTG.......................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................AGGCTGGGGAGCCGGGCTTA........ | 20 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TGGCGGTCGGGCTCTGCGTGTCCGG....................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................CGAAGTTGTGTTTCCCGAGTGCACGGGC.................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AGGCTGGCACGAAGTTGTG...................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AGGCTGGCACGAAGTTG........................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................ACGAAGTTGTGTTTCCCGAGTGCA......................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................GCGGTCGGGCTCTGCGTGTCCGGaa..................................................................................................................................................................................................... | 25 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGAGAAGGCTGGCACGA............................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GTGTCCGGTGAGGGAGGGGAGAAGG...................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................AGGGGAGAAGGCTGGCAC............................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGGAGTCCGAGTGTCCCCCGCCGTT...................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TCCGGTGAGGGAGGGGAGAAGGC..................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGAGAAGGCTGGCACGAAG........................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................GGGAGAAGGCTGGCACGAAGTTG........................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................CGGGCGCGTGCGGGGCTC....................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGAGAAGGCTGGCACGAAGTT......................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................TCCTCGCTGGAGTCCGAGTGTCCCC............................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGGAGTCCGAGTGTCCCCCGCCGT....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGGAGTCCGAGTGTCCCC............................................................................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TCGCTGGAGTCCGAGTGTCCCCCGCC......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................GGGAGAAGGCTGGCACGAAGTTGTG...................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CCGGTGAGGGAGGGGAGAAGGCTG................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GTGTCCGGTGAGGGAGGGGAGAAGGCTGGC................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................GGGCTCTGCGTGTCCGGT...................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GTGTCCGGTGAGGGAGGG............................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................GCCGGGCTTACTCAGCG. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................AAGGCTGGCACGAAGTTGTG...................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CCCGCGGCGAGGCTGGGGAGCCGGGCTTAa....... | 30 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GTGCACGGGCGCGTGCGGGGCTCGGG.................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................GGGAGGGGAGAAGGCTGGC................................................................................................................................................................................. | 19 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GAGGGGAGAAGGCTGGCA................................................................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................GAGGGAGGGGAGAAGGCTG................................................................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ....................................................GAGGGAGGGGAGAAGGCTGtc................................................................................................................................................................................. | 21 | tc | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................GGAGGGGAGAAGGCTGGC................................................................................................................................................................................. | 18 | 3 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGCGGGGCTCGGGAGG................................................................................................................................. | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - |
| GGCTCCGTGCGCCGGAAGCGAGCCGCTGGCGGTCGGGCTCTGCGTGTCCGGTGAGGGAGGGGAGAAGGCTGGCACGAAGTTGTGTTTCCCGAGTGCACGGGCGCGTGCGGGGCTCGGGAGGAGTTGTGCGGCTCCTCGCTGGAGTCCGAGTGTCCCCCGCCGTTCCCTGTGCTGCTCGCGAGCGCCACTTTGGGGGAAGAGCAGCCTCTTGCCCCCGCGGCGAGGCTGGGGAGCCGGGCTTACTCAGCGC ...................................................................(((.((((((.......(((((((((((((((....)))))...))))))))))..)))))))))...................................................................................................................... ......................................................55...........................................................................132.................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................GCGGCGAGGCTGGGGAGCCGGtg............. | 23 | tg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |