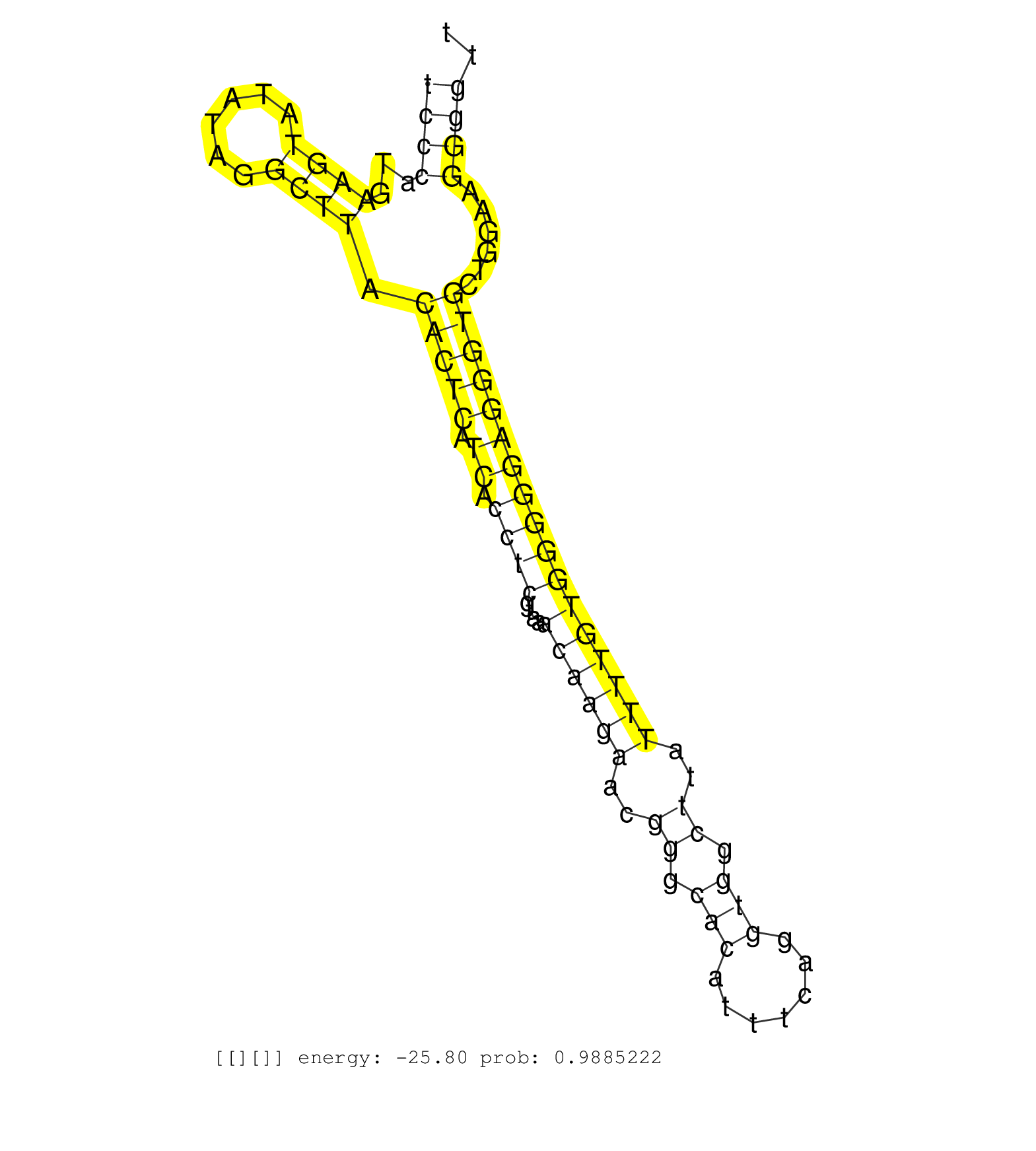
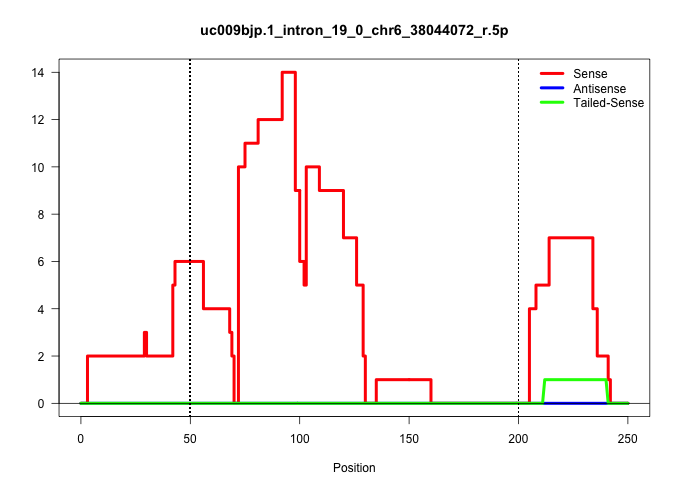
| Gene: Atp6v0a4 | ID: uc009bjp.1_intron_19_0_chr6_38044072_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| CACTGAACGTCGTCATTTAGTCCAGCCCATTGCAGGCATTCCTGGAACAGGTGAGAGGGTTTCTACTTCCCATGAAGTATATAGGCTTACACTCATCACCTCGTAAACAAGAACGGGCACATTTCAGGTGGCTTATTTTGTGGGGGAGGGTGCTGGAAGGGGTTGGTGGTGATTGTGGGTGTCTAAACTGGAATTCTTTTGTCCATGTTAAAAAGACTTGAAAACTGAAACGTCACGGTACAGTGTTAAT ...................................................................((((...((((......)))).(((((.((.((((....((((((..((.(((.......))).))..)))))))))))))))))......))))........................................................................................ ...................................................................68..............................................................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................TGAAGTATATAGGCTTACACTCATCA........................................................................................................................................................ | 26 | 1 | 5.00 | 5.00 | 2.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................TGAAGTATATAGGCTTACACTCATCACC...................................................................................................................................................... | 28 | 1 | 5.00 | 5.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGTTAAAAAGACTTGAAAACTGAAACGTC................ | 29 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................TAAACAAGAACGGGCACATTTCAGGT......................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TTGCAGGCATTCCTGGAACAGGTGAGA.................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| .......................................................................................................TAAACAAGAACGGGCACATTTCAGGTG........................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TCATCACCTCGTAAACAAGAACGGGCAC.................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TCGTAAACAAGAACGGGCACATTTCA............................................................................................................................ | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................................TAAAAAGACTTGAAAACTGAAACGTCAC.............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................TGGAACAGGTGAGAGGGTTTCTACTTC..................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TAAAAAGACTTGAAAACTGAAACGTCACG............. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................GGAACAGGTGAGAGGGTTTCTACTTCC.................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AGTATATAGGCTTACACTCATCACCTC.................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GACTTGAAAACTGAAACGTCACGGTACA........ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TAGGCTTACACTCATCACCTCGTAAACA............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................TGGAACAGGTGAGAGGGTTTCTACTT...................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGTTAAAAAGACTTGAAAACTGAAACGTCAC.............. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................AAGACTTGAAAACTGAAACGTCACGGTAt......... | 29 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TTTTGTGGGGGAGGGTGCTGGAAGG.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...TGAACGTCGTCATTTAGTCCAGCCCAT............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGGAACAGGTGAGAGGGTTTCTACTTCC.................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GACTTGAAAACTGAAACGTCACGGTAC......... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGAACGTCGTCATTTAGTCCAGCCCA............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACTGAACGTCGTCATTTAGTCCAGCCCATTGCAGGCATTCCTGGAACAGGTGAGAGGGTTTCTACTTCCCATGAAGTATATAGGCTTACACTCATCACCTCGTAAACAAGAACGGGCACATTTCAGGTGGCTTATTTTGTGGGGGAGGGTGCTGGAAGGGGTTGGTGGTGATTGTGGGTGTCTAAACTGGAATTCTTTTGTCCATGTTAAAAAGACTTGAAAACTGAAACGTCACGGTACAGTGTTAAT ...................................................................((((...((((......)))).(((((.((.((((....((((((..((.(((.......))).))..)))))))))))))))))......))))........................................................................................ ...................................................................68..............................................................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|