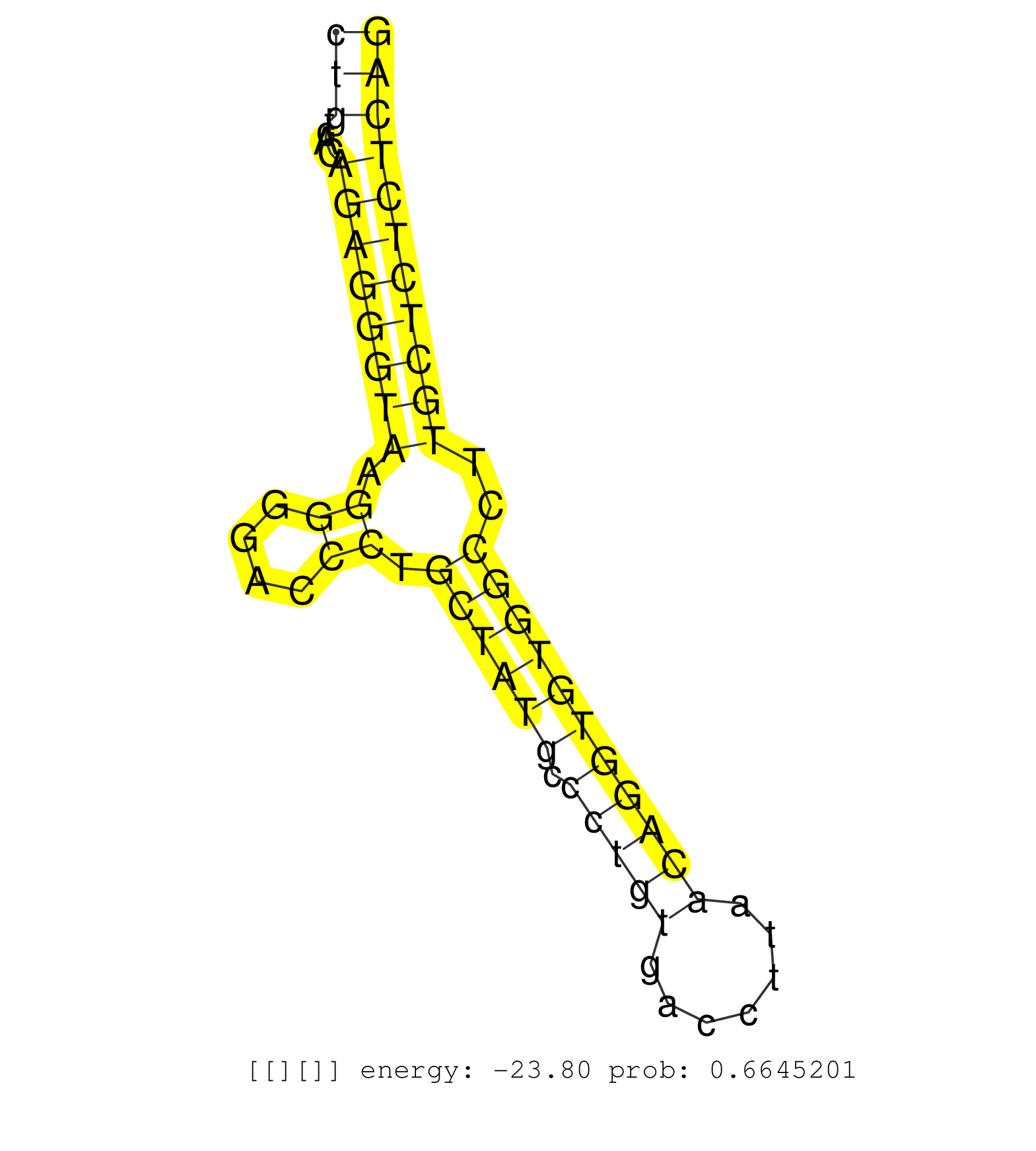
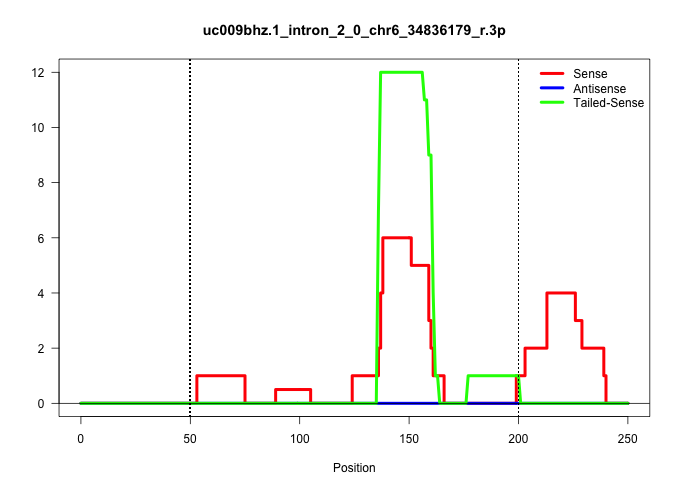
| Gene: Wdr91 | ID: uc009bhz.1_intron_2_0_chr6_34836179_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(15) TESTES |
| AGGAACCCCAGGTGAGCTCTGTCCCCAAGGATCCCCAGTCCTCCCTTTGACCATATCAGGAGGCTGCTCAGAAAAGGTGACCTGGAAGGGTCAGCACTTTGTGGCCCAGGAGGCCCCTGGCCACTAGAGCAGCTGTCACAGAGGGTAAGGGGACCCTGCTATGCCCTGTGACCTTAACAGGTGTGGCCTTGCTCTCTCAGACATGCAGCAGCACGAGTGTGCTATGAGCTGGAAGGCCCACTGCGGCGAG ....................................................................................................................................(((....((((((((.((....)).((((((.(((((.......)))))))))))..))))))))))).................................................. ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................ACAGAGGGTAAGGGGACCCTGC........................................................................................... | 22 | 1 | 8.00 | 8.00 | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ACAGAGGGTAAGGGGACCCTGtaaa........................................................................................ | 25 | taaa | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GACATGCAGCAGCACGAGTGTGCTATG........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................ACAGGTGTGGCCTTGCTCTCT..................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CACAGAGGGTAAGGGGACCCTGCaa......................................................................................... | 25 | aa | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CACAGAGGGTAAGGGGACCCTGaaga........................................................................................ | 26 | aaga | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CGAGTGTGCTATGAGCTGGAAGGCCCA.......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................TCAGGAGGCTGCTCgttt................................................................................................................................................................................. | 18 | gttt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CACAGAGGGTAAGGGGACCCTGaat......................................................................................... | 25 | aat | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TAGAGCAGCTGTCACAGAGGGTAAGGG................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ACAGAGGGTAAGGGGACCCTGaaa......................................................................................... | 24 | aaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CACAGAGGGTAAGGGGACCCc............................................................................................. | 21 | c | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CACAGAGGGTAAGGGGACCCTGCat......................................................................................... | 25 | at | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CAGAGGGTAAGGGGACCCTGC........................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................CAGAGGGTAAGGGGACCCTGCTA......................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CACAGAGGGTAAGGGGACCCTGCTAatt...................................................................................... | 28 | att | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGTGTGGCCTTGCTCTCTCAGt................................................. | 24 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................CAGAGGGTAAGGGGACCCTGCa.......................................................................................... | 22 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................CACAGAGGGTAAGGGGACCCTGt........................................................................................... | 23 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CACAGAGGGTAAGGGGACCCTGC........................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................CACAGAGGGTAAGGGGACCCTGtaag........................................................................................ | 26 | taag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................................................................TGCAGCAGCACGAGTGTGCTATGAGC..................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ACAGAGGGTAAGGGGACCCTGCTATGCCC.................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TATCAGGAGGCTGCTCAGAAAA............................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................ACAGAGGGTAAGGGGACCCTGCat......................................................................................... | 24 | at | 1.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ACAGAGGGTAAGGGGACCCTGCT.......................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ACAGAGGGTAAGGGGACCCTGa........................................................................................... | 22 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CGAGTGTGCTATGAGCTGGAAGGCCC........... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................GTCAGCACTTTGTGGC................................................................................................................................................. | 16 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGAACCCCAGGTGAGCTCTGTCCCCAAGGATCCCCAGTCCTCCCTTTGACCATATCAGGAGGCTGCTCAGAAAAGGTGACCTGGAAGGGTCAGCACTTTGTGGCCCAGGAGGCCCCTGGCCACTAGAGCAGCTGTCACAGAGGGTAAGGGGACCCTGCTATGCCCTGTGACCTTAACAGGTGTGGCCTTGCTCTCTCAGACATGCAGCAGCACGAGTGTGCTATGAGCTGGAAGGCCCACTGCGGCGAG ....................................................................................................................................(((....((((((((.((....)).((((((.(((((.......)))))))))))..))))))))))).................................................. ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|