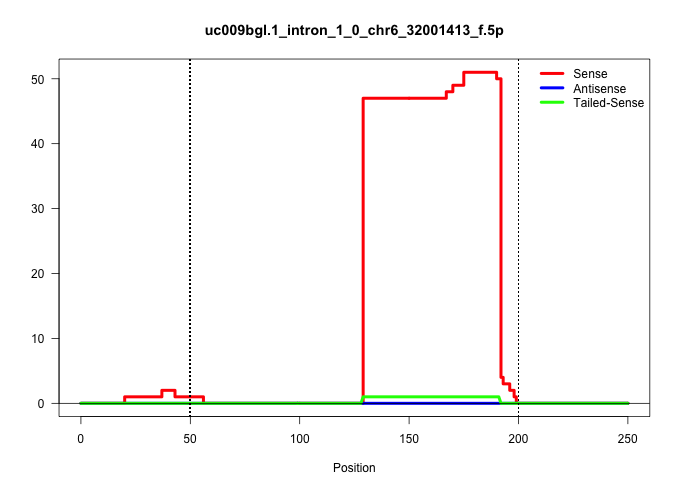
| Gene: 1700012A03Rik | ID: uc009bgl.1_intron_1_0_chr6_32001413_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(8) TESTES |
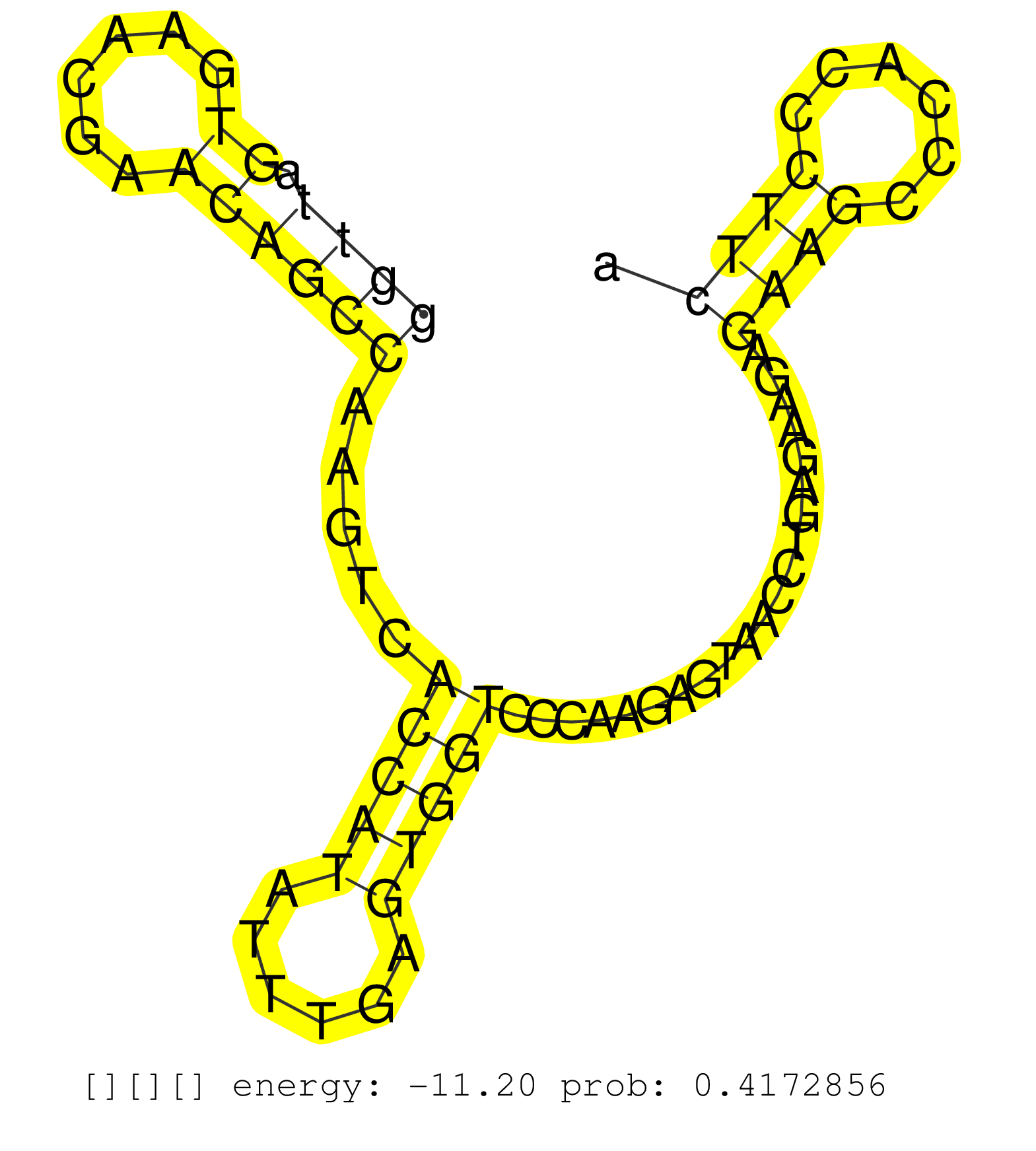
| AAGTAGAACTATGAAAACCAAACCCAAAGCAACTTCCAGGAGACTCCAAAGTAAGCCAAGCATGAGTGATGTGAGTGGTCGGTCAGGTTTACTTATGGCTGTGGAGCCAGGAAGCTTAATTTGAGGTTAGTGAACGAACAGCCAAGTCACCATATTTGAGTGGTCCCAAGAGTAACCTGAGAAGAGAAGCCCACCCTTCATCTTGCTTCACCTCCTGGCTCCTTTGTCACCTGACACTTCGCTTCCTCAT ............................................................................................................................((((.((......)))))).....(((((......))))).....................((((......))))................................................... ............................................................................................................................125........................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................GTGAACGAACAGCCAAGTCACCATATTTGAGTGGTCCCAAGAGTAACCTGAG..................................................................... | 52 | 1 | 48.00 | 48.00 | 48.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................TTAGTGAACGAACAGCCAAG........................................................................................................ | 20 | 1 | 9.00 | 9.00 | - | 9.00 | - | - | - | - | - | - |
| ................................................................................................................................................................TGGTCCCAAGAGTAACCTGAGAAGAGAAGC............................................................ | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - |
| .....................................AGGAGACTCCAAAGTAAGC.................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................CCTGAGAAGAGAAGCCCACCCTT.................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................................AGTAACCTGAGAAGAGAAGC............................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................GGTTTACTTATGGCTGTGGA................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................AAGAGTAACCTGAGAAGAGAAGCCCACCCTTC................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ....................AACCCAAAGCAACTTCCAGGAGA............................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................CCTGAGAAGAGAAGCCCACCC...................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................AGGTTAGTGAACGAACAGC............................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| AAGTAGAACTATGAAAACCAAACCCAAAGCAACTTCCAGGAGACTCCAAAGTAAGCCAAGCATGAGTGATGTGAGTGGTCGGTCAGGTTTACTTATGGCTGTGGAGCCAGGAAGCTTAATTTGAGGTTAGTGAACGAACAGCCAAGTCACCATATTTGAGTGGTCCCAAGAGTAACCTGAGAAGAGAAGCCCACCCTTCATCTTGCTTCACCTCCTGGCTCCTTTGTCACCTGACACTTCGCTTCCTCAT ............................................................................................................................((((.((......)))))).....(((((......))))).....................((((......))))................................................... ............................................................................................................................125........................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................AGCCCACCCTTCATaatt................................................. | 18 | aatt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - |