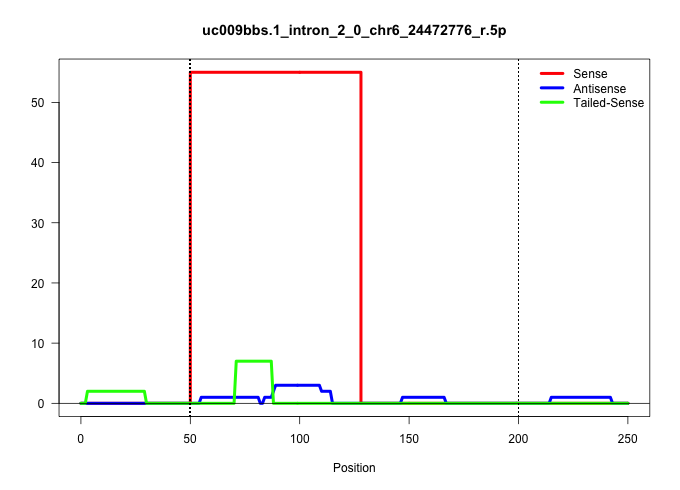
| Gene: Ndufa5 | ID: uc009bbs.1_intron_2_0_chr6_24472776_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(10) TESTES |
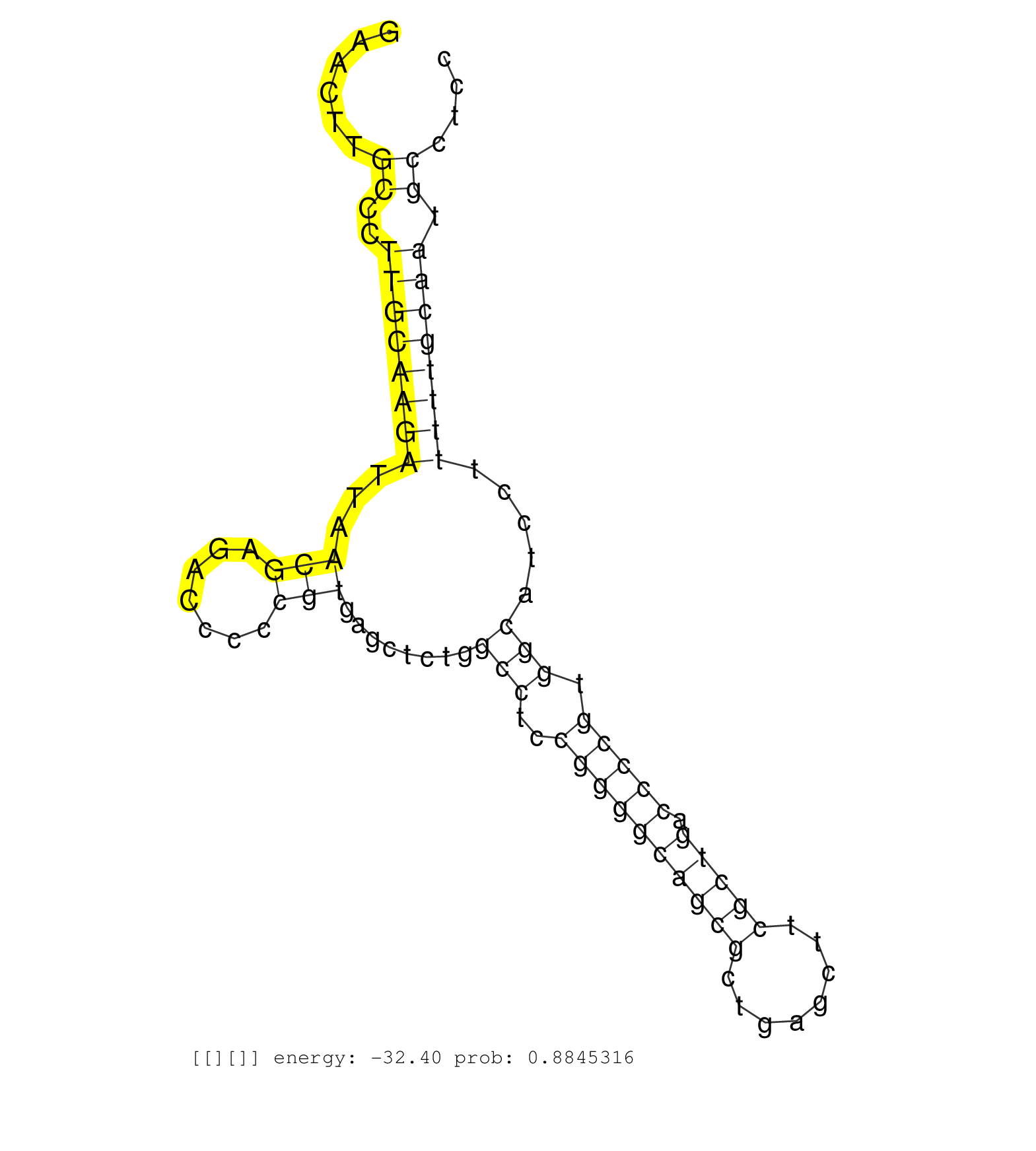
| CACAGACAACTGGCCTGGTGGGGTTGGCGGTGTGCGACACTCCACACGAGGTAAGTGCGACTTTTTGTGTCGTCCTTCTCAAGCAAGACTGTGATCGACCGAACTTGCCCTTGCAAGATTAACGAGACCCCCGTGAGCTCTGGCCTCCGGGGCAGCGCTGAGCTTCGCTGACCCCGTGGCATCCTTTTTGCAATGCCTCCTTTTGCCTTCGCCGTGACCTCTTAATTAAGACGCGTTTCTTCACGGCATT ..........................................................................................................((..((((((((...(((.......)))........(((..((((((((((........))))).))))).))).....)))))))).))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTGCGACTTTTTGTGTCGTCCTTCTCAAGCAAGACTGTGATCGACCGA.................................................................................................................................................... | 52 | 1 | 55.00 | 55.00 | 55.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................ACTGTGATCGACCGAACTTGCC............................................................................................................................................. | 22 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - |
| ...AGACAACTGGCCTGGTGGGGTTGGCGt............................................................................................................................................................................................................................ | 27 | t | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - |
| ............GCCTGGTGGGGTTGGCGGTGTGC....................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ...AGACAACTGGCCTGGTGGGGTTGGCGGTGTGCGAC.................................................................................................................................................................................................................... | 35 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| CACAGACAACTGGCCTGGTGGGGTTGGCGGTGTGCGACACTCCACACGAGGTAAGTGCGACTTTTTGTGTCGTCCTTCTCAAGCAAGACTGTGATCGACCGAACTTGCCCTTGCAAGATTAACGAGACCCCCGTGAGCTCTGGCCTCCGGGGCAGCGCTGAGCTTCGCTGACCCCGTGGCATCCTTTTTGCAATGCCTCCTTTTGCCTTCGCCGTGACCTCTTAATTAAGACGCGTTTCTTCACGGCATT ..........................................................................................................((..((((((((...(((.......)))........(((..((((((((((........))))).))))).))).....)))))))).))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................CGGGGCAGCGCTGAGCTTCG................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................TGCGACTTTTTGTGTCGTCCTTCTCAA........................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................AAGACTGTGATCGACCGAACTTGCCC............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................CTGTGATCGACCGAACTTGCCCTTGCA....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................TGTGATCGACCGAACTTGCCCTTGCA....................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................................................................................................GACCTCTTAATTAAGACGCGTTTCTTCA....... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................CGCTGACCCCGTGgcga........................................................................ | 17 | gcga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - |