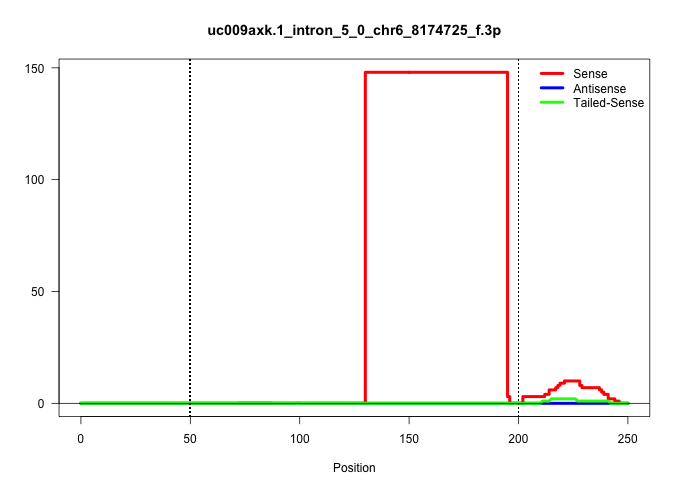
| Gene: BC020002 | ID: uc009axk.1_intron_5_0_chr6_8174725_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| AATGGCCTGCGTTTTTATGTCTATCTAACTGAATTAGCTAGGTCTGTCTGCATTTGAAGGACAAAGCTTCACTGTTAAGGACAGTGGCCTTCCGAGGCGTCTTTAGCAATTGATGACTCTTCTAGCATCTGAACAGTACCTTTGGAACACATTTCAAAATAAACTTATAAATAAATGGCGTGTATAATTTTCCCCCCCAGTATGAAAACAAAGTTGCTGTACGTGACAGAGTGGCATTCGCTTGTAAATT ....................................................................(((.((((((((((....((((....))))))))))))))..)))..(((.(((........))).)))...(((((((....)))))))............................................................................................ ...................................................................68..................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................GAACAGTACCTTTGGAACACATTTCAAAATAAACTTATAAATAAATGGCGTG.................................................................... | 52 | 1 | 148.00 | 148.00 | 148.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGAAAACAAAGTTGCTGTACGTGACA...................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................................................GCTGTACGTGACAGAGTGGCATTCttt........ | 27 | ttt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GTACGTGACAGAGTGGCATT............ | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AGTTGCTGTACGccct....................... | 16 | ccct | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGCTGTACGTGACAGAGTGGCAT............. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGTACGTGACAGAGTGGCATTCGCTTG...... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................................................................................................TACGTGACAGAGTGGCATTCGCTTGTA.... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGCTGTACGTGACAGAGTGGCATTCGC......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................................................GTTGCTGTACGTGACAGAGTGGCATTC........... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................................................CGTGACAGAGTGGCATTCGC......... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGTACGTGACAGAGTGGCATTCGCTTGTA.... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................................................TGAAAACAAAGTTGCTGTACGTGACAG..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................TGTTAAGGACAGTGG................................................................................................................................................................... | 15 | 13 | 0.08 | 0.08 | - | - | 0.08 | - | - | - | - | - | - | - | - |
| AATGGCCTGCGTTTTTATGTCTATCTAACTGAATTAGCTAGGTCTGTCTGCATTTGAAGGACAAAGCTTCACTGTTAAGGACAGTGGCCTTCCGAGGCGTCTTTAGCAATTGATGACTCTTCTAGCATCTGAACAGTACCTTTGGAACACATTTCAAAATAAACTTATAAATAAATGGCGTGTATAATTTTCCCCCCCAGTATGAAAACAAAGTTGCTGTACGTGACAGAGTGGCATTCGCTTGTAAATT ....................................................................(((.((((((((((....((((....))))))))))))))..)))..(((.(((........))).)))...(((((((....)))))))............................................................................................ ...................................................................68..................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|