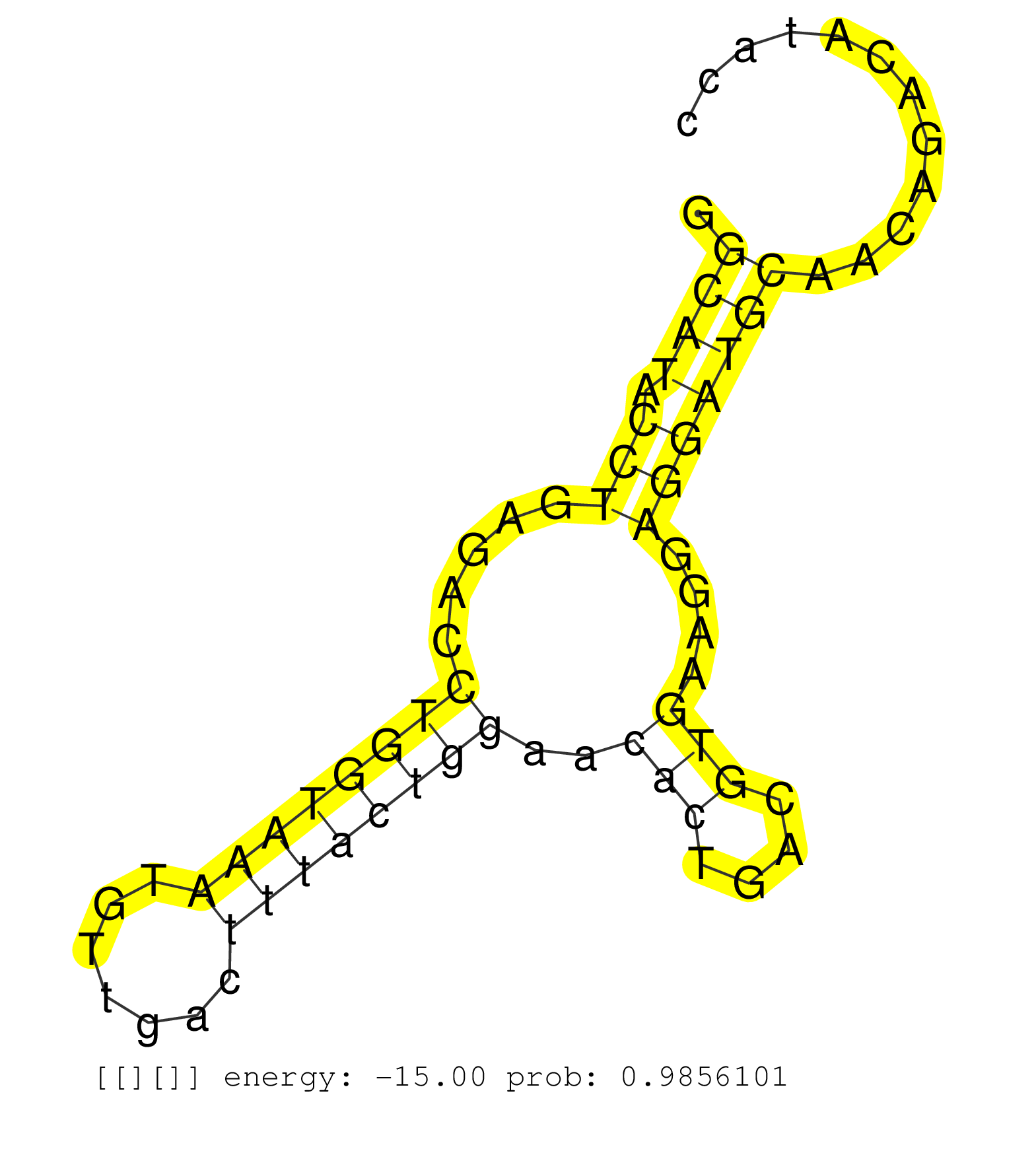
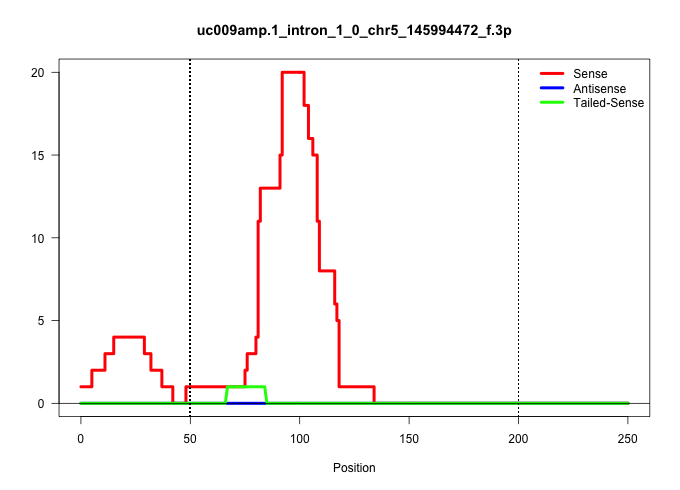
| Gene: Zfp655 | ID: uc009amp.1_intron_1_0_chr5_145994472_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(9) TESTES |
| TATTACAGTCCTCAGTCTGTGTAAGTAGGAGCAGCTCCCAAGGAGGAGTGGGCATACCTGAGACCTGGTAAATGTTGACTTTACTGGAACACTGACGTGAAGGAGGATGCAACAGACATACCTTGGGCAGGAGAAGTGTTCTGAAAAGGGAGCCCTGTGGAGGGGCTGCTCTCACTCTTGGCTCCTTTCTCCTCCCTCAGCTCCACCTGTTCCCCAGGTGCCTGCTCTTCCCCATGAGGGAAGCCCAGGA ...................................................((((.(((.....((((((((.......))))))))..(((....)))....)))))))............................................................................................................................................ ..................................................51.....................................................................122.............................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................TGACGTGAAGGAGGATGCAACAGACA.................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - |
| .................................................................................TACTGGAACACTGACGTGAAGGAGGAT.............................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - |
| .................................................................................TACTGGAACACTGACGTGAAGGAGGATG............................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | 1.00 | 2.00 | - | - | - | - | - |
| ...........................................................................TGACTTTACTGGAACACTGACGTGAAGGA.................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ................................................................................TTACTGGAACACTGACGTGAAGGAGG................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................GACTTTACTGGAACACTGACGTGAAG.................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................CTGACGTGAAGGAGGATGCAACAGA...................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................TGACGTGAAGGAGGATGCAACAGA...................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................TACTGGAACACTGACGTGAAGGAGGA............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................ACTGGAACACTGACGTGAAG.................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................TGGAACACTGACGTGAAGGAGGATGC............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ................................................TGGGCATACCTGAGACCTGGTAAATGT............................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................CTGACGTGAAGGAGGATGCAACAGAC..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................ACTGGAACACTGACGTGAAGGAGGAT.............................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................TGCAACAGACATACCTTGGGCAGGAGA.................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ...............TCTGTGTAAGTAGGAGCAGCTCCCAAG................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................GTAAATGTTGACTTgtgc..................................................................................................................................................................... | 18 | gtgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |
| .....CAGTCCTCAGTCTGTGTAAGTAGGAGC.......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ...........TCAGTCTGTGTAAGTAGGAGCAGCTC..................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| TATTACAGTCCTCAGTCTGTGTAAGTAGGAGCAGCTCCCAAGGAGGAGTGGGCATACCTGAGACCTGGTAAATGTTGACTTTACTGGAACACTGACGTGAAGGAGGATGCAACAGACATACCTTGGGCAGGAGAAGTGTTCTGAAAAGGGAGCCCTGTGGAGGGGCTGCTCTCACTCTTGGCTCCTTTCTCCTCCCTCAGCTCCACCTGTTCCCCAGGTGCCTGCTCTTCCCCATGAGGGAAGCCCAGGA ...................................................((((.(((.....((((((((.......))))))))..(((....)))....)))))))............................................................................................................................................ ..................................................51.....................................................................122.............................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|