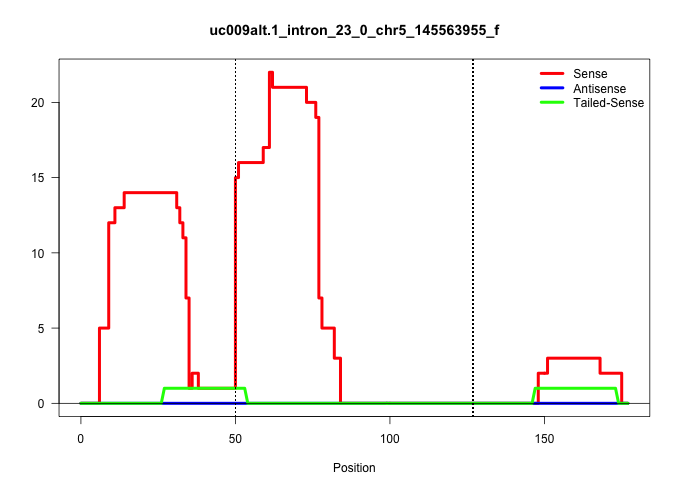
| Gene: Trrap | ID: uc009alt.1_intron_23_0_chr5_145563955_f | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| TGGCCGTGATATTTGACGTGGCGAGCATCATCCTGGGCTCAAAAGAGAGGGTAAGGACGTCTGAACAGTTTGCAGGCGCTTGGTCAGAAAGGCAGCACCGACTTATTCCTGGCTTGTTTCTCTCCAGGCATGCCAGCTGCCCCTCTTCTCCTACATTGTGGAGCGGTTGTGCGCCTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGGACGTCTGAACAGTTTGCAGGC.................................................................................................... | 27 | 1 | 11.00 | 11.00 | 4.00 | - | 1.00 | - | 1.00 | - | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - |
| .........TATTTGACGTGGCGAGCATCATCCTG.............................................................................................................................................. | 26 | 1 | 5.00 | 5.00 | - | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - |
| .............................................................TGAACAGTTTGCAGGCGCTTGGT............................................................................................. | 23 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGATATTTGACGTGGCGAGCATCATCCT............................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCCTACATTGTGGAGCGGTTGTGCGCC.. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................TGAACAGTTTGCAGGCGCTTG............................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TACATTGTGGAGCGGTT......... | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TATTTGACGTGGCGAGCATCAT.................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGATATTTGACGTGGCGAGCATCATC................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACGTCTGAACAGTTTGCAGGCG................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................TCTGAACAGTTTGCAGGCG................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TTTGACGTGGCGAGCATCATCCTGGGC........................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACGTCTGAACAGTTTGCAGG..................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................GCTCAAAAGAGAGGGTAAGGACGTCT................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............GACGTGGCGAGCATCATCCTG.............................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................CTCCTACATTGTGGAGCGGTTGTGCGt... | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACGTCTGAACAGTTTGC........................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................TAAGGACGTCTGAACAGTTTGCAGGC.................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......TGATATTTGACGTGGCGAGCATCATCC................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TCATCCTGGGCTCAAAAGAGAGGGcat........................................................................................................................... | 27 | cat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........TATTTGACGTGGCGAGCATCATCCT............................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| TGGCCGTGATATTTGACGTGGCGAGCATCATCCTGGGCTCAAAAGAGAGGGTAAGGACGTCTGAACAGTTTGCAGGCGCTTGGTCAGAAAGGCAGCACCGACTTATTCCTGGCTTGTTTCTCTCCAGGCATGCCAGCTGCCCCTCTTCTCCTACATTGTGGAGCGGTTGTGCGCCTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|