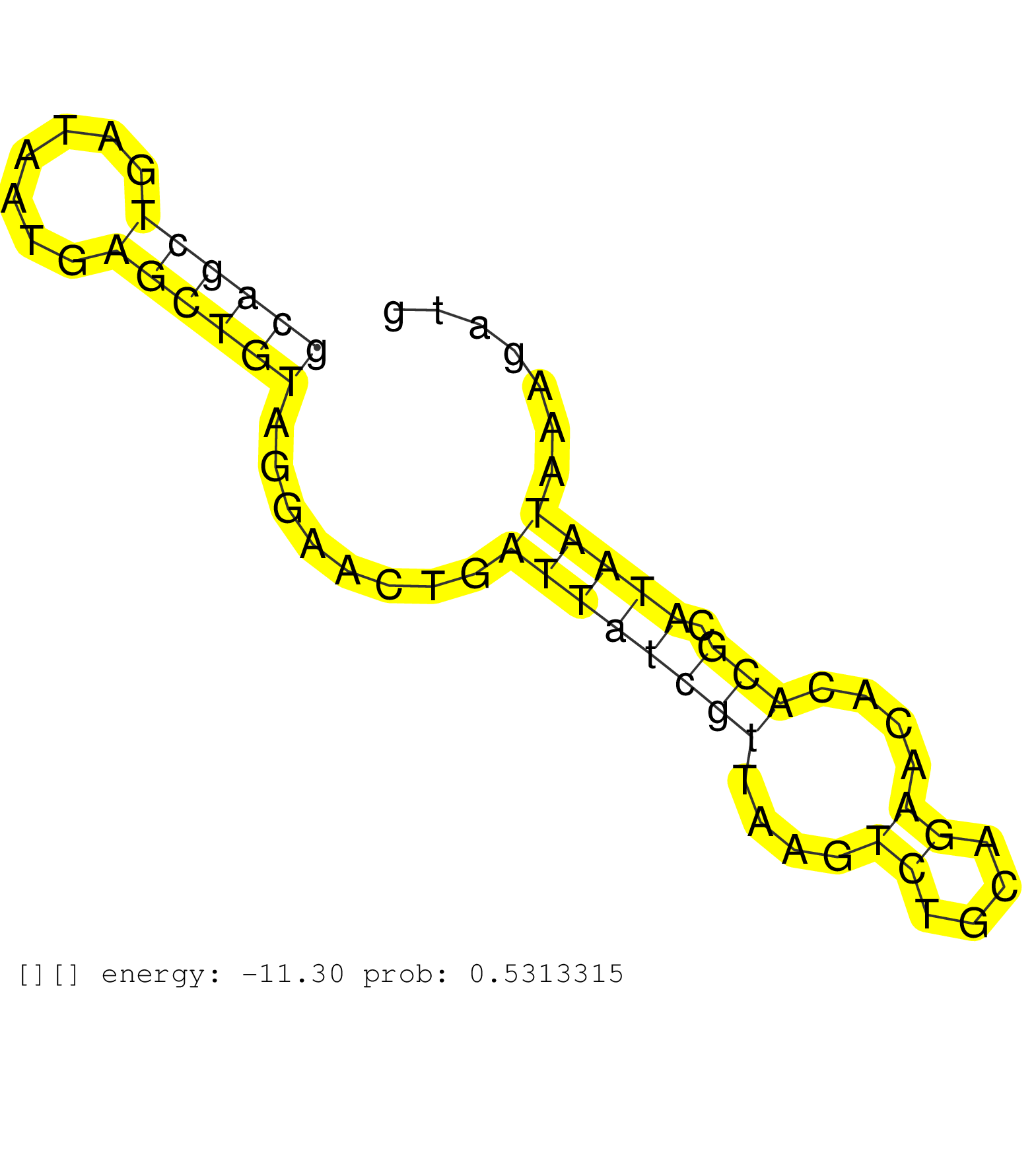
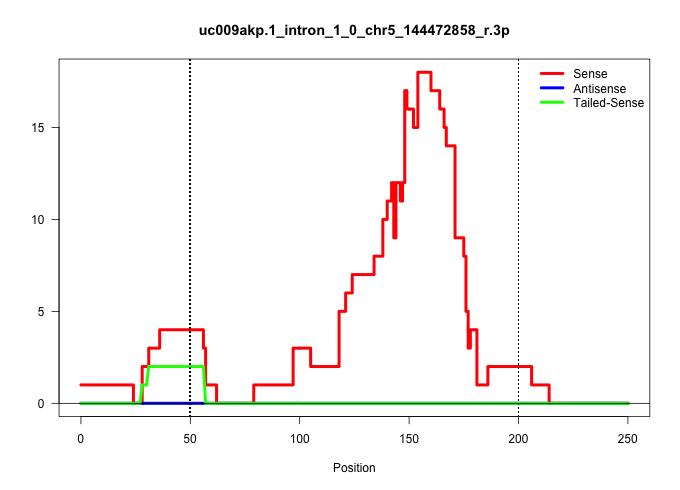
| Gene: Usp42 | ID: uc009akp.1_intron_1_0_chr5_144472858_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| TGTGCAGCTCTGTGCTGGGGCTGAGACCTGGTCCCTCTTGTGTGGCTCTTGACGGTCCTGTGCCGGTGCTTTGTGGCACTGCCAGCTCATGCTGCTATCCGAGTGATTTTTCAGCAGCTGATAATGAGCTGTAGGAACTGATTATCGTTAAGTCTGCAGAACACACGCATAATAAAGATGTTCCTTACTTTGCCTTCTAGGTGACTAAACAGCCTTGGCCTTACAACAACAAAATTACGGTAAGCAATTG .................................................................................................................((((((.......))))))........((((((((....((....))....))).)))))............................................................................. .................................................................................................................114...............................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................TAAGTCTGCAGAACACACGCATAATAAA.......................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TGCAGAACACACGCATAATAAAGATGT..................................................................... | 27 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGATAATGAGCTGTAGGAACTGATT........................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................TCGTTAAGTCTGCAGAACACACGCATA............................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - |
| .................................................................................................TCCGAGTGATTTTTCAGCAGCTGATA............................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAAGTCTGCAGAACACACGCATAATAAAG......................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TAATGAGCTGTAGGAACTGATTATC........................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................TCCCTCTTGTGTGGCTCTTGACGGTC................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGTTCCTTACTTTGCCTTCTAGGTGACT............................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................TGGTCCCTCTTGTGTGGCTCTTGACGGT.................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CTTGTGTGGCTCTTGACGGTCCTGTG............................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ATGAGCTGTAGGAACTGATTATCGTT..................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CGTTAAGTCTGCAGAACACACGCATA............................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGATTATCGTTAAGTCTGCAGAACACAC.................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................TGGTCCCTCTTGTGTGGCTCTTGACGGTC................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TTAAGTCTGCAGAACACACGCATAATAA........................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ATGAGCTGTAGGAACTGATTAT......................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGATTATCGTTAAGTCTGCAGAACAC...................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TGGTCCCTCTTGTGTGGCTCTTGACGGga................................................................................................................................................................................................. | 29 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................ACTTTGCCTTCTAGGTGACTAAACAGCC.................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................TCCCTCTTGTGTGGCTCTTGACGGTt................................................................................................................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TGCCAGCTCATGCTGCTATCCGAGTG................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................GAACTGATTATCGTTAAGTCTGCAGA.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ATTATCGTTAAGTCTGCAGAACACACG................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TATCGTTAAGTCTGCAGAACACACGCATA............................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGAGCTGTAGGAACTGATTATCGTTAAG.................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| TGTGCAGCTCTGTGCTGGGGCTGAGACCTGGTCCCTCTTGTGTGGCTCTTGACGGTCCTGTGCCGGTGCTTTGTGGCACTGCCAGCTCATGCTGCTATCCGAGTGATTTTTCAGCAGCTGATAATGAGCTGTAGGAACTGATTATCGTTAAGTCTGCAGAACACACGCATAATAAAGATGTTCCTTACTTTGCCTTCTAGGTGACTAAACAGCCTTGGCCTTACAACAACAAAATTACGGTAAGCAATTG .................................................................................................................((((((.......))))))........((((((((....((....))....))).)))))............................................................................. .................................................................................................................114...............................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|