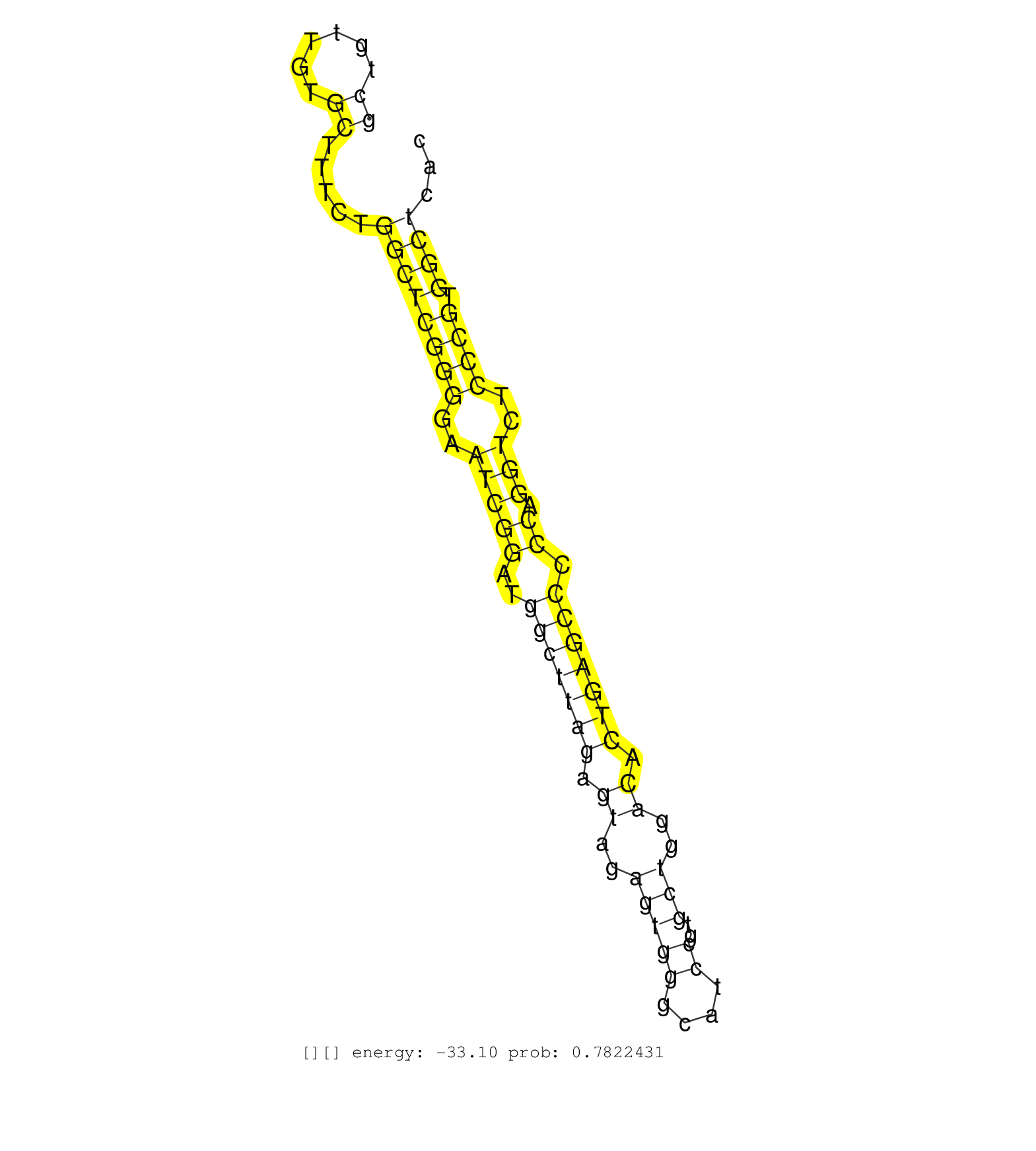
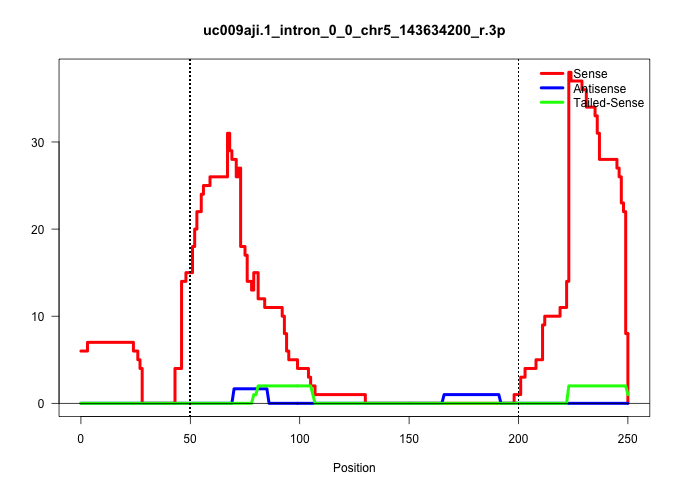
| Gene: Fbxl18 | ID: uc009aji.1_intron_0_0_chr5_143634200_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(6) PIWI.ip |
(3) PIWI.mut |
(1) TDRD1.ip |
(26) TESTES |
| TAGGTGCTAAAACCCTGCAGTCCGGCCTCAGGCCCCGGACCGCTGTTGTGCTTTCTGGCTCGGGGAATCGGATGGCTTAGAGTAGAGTGGGCATCCGTGCTGGACACTGAGCCCCCAGGTCTCCCGTGGCTCACGGAGCACTGTGCCTTCCCAGCTGTGTCCTCAGCTCACACCCCGTTAACGGCTGACCTCTGGTTCAGTTTCCAGGCCGAGAGGCCCGCGTTGAACGTCGTCATCTTCCCCCTGCTCC .........................................((......)).....((((((((..(((((..(((((((.((..(((((....))..)))..)).))))))).)).)))..)))).))))....................................................................................................................... .........................................42..........................................................................................134.................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................TGAACGTCGTCATCTTCCCCCTGCTC. | 26 | 1 | 13.00 | 13.00 | 3.00 | - | 3.00 | - | 1.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................TTGAACGTCGTCATCTTCCCCCTGC... | 25 | 1 | 9.00 | 9.00 | - | - | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGAACGTCGTCATCTTCCCCCTGCTCC | 27 | 1 | 8.00 | 8.00 | 1.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................TGTGCTTTCTGGCTCGGGGAATCGGAT................................................................................................................................................................................. | 27 | 1 | 8.00 | 8.00 | - | 4.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................TCGGATGGCTTAGAGTAGAGTGGGC.............................................................................................................................................................. | 25 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGAACGTCGTCATCTTCCCCCTGC... | 24 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................TCGGATGGCTTAGAGTAGAGTGGGCA............................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................................................AGAGGCCCGCGTTGAACGTCGTCATC............. | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TTCTGGCTCGGGGAATCGGATGGC.............................................................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AGAGGCCCGCGTTGAACGTCGTCAT.............. | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................TCGGATGGCTTAGAGTAGAGTGGGCAT............................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TGTGCTTTCTGGCTCGGGGAATCGG................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TGTTGTGCTTTCTGGCTCGGGGAAT...................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTGAACGTCGTCATCTTCCCCCTGCTC. | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGGCTCGGGGAATCGGATGGCTTAGA......................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TTTCTGGCTCGGGGAATCGGATGGCTT............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................CACTGAGCCCCCAGGTCTCCCGTGGC........................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TCGGATGGCTTAGAGTAGAGTGGGCATC........................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGTTTCCAGGCCGAGAGGCCCGCGTT.......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTCCAGGCCGAGAGGCCCGCGTTGAACGTC................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................GAGTAGAGTGGGCATCCGTGCTGGA.................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................GAGTAGAGTGGGCATCCGTGCTGGACAC............................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCGGGGAATCGGATGGCTTAGAGTA...................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................GAGTAGAGTGGGCATCCGTGCTGGAC................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TCTGGCTCGGGGAATCGGATGGCTTA........................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TCTGGCTCGGGGAATCGGATGG............................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TGTTGTGCTTTCTGGCTCGGGGA........................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GGCTCGGGGAATCGGATGGCTTAGA......................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TTTCTGGCTCGGGGAATCGGATGGC.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGAACGTCGTCATCTTCCCCCTGCTCCg | 28 | g | 1.00 | 8.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...GTGCTAAAACCCTGCAGTCCG.................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................................................................................CCGAGAGGCCCGCGTTGAACGTCGTCA............... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TAGAGTAGAGTGGGCATCCGTGCTGGA.................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGGCTTAGAGTAGAGTGGGCATCCGTG....................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTGAACGTCGTCATCTTCCCCCTG.... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GTAGAGTGGGCATCCGTGCTGGACAa............................................................................................................................................... | 26 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................TGCTTTCTGGCTCGGGGAATCGGAT................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................GAGGCCCGCGTTGAACGTCGTCATC............. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGAACGTCGTCATCTTCCCCCTGCT.. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TTTCTGGCTCGGGGAATCGGATGGCT............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................CCAGGCCGAGAGGCCCGCGTTGAACGTC................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATCGGATGGCTTAGAGTAGAGTGGGC.............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TGTTGTGCTTTCTGGCTCGGGGAATC..................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GCGTTGAACGTCGTCATCTTCCCCCT..... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTCCAGGCCGAGAGGCCCGCGTTGAACG..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................GAGTAGAGTGGGCATCCGTGCTGGACt................................................................................................................................................ | 27 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGAACGTCGTCATCTTCCCCCTGCTCa | 27 | a | 1.00 | 13.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TAGGTGCTAAAACCCTGCAGTCCGGCCTCAGGCCCCGGACCGCTGTTGTGCTTTCTGGCTCGGGGAATCGGATGGCTTAGAGTAGAGTGGGCATCCGTGCTGGACACTGAGCCCCCAGGTCTCCCGTGGCTCACGGAGCACTGTGCCTTCCCAGCTGTGTCCTCAGCTCACACCCCGTTAACGGCTGACCTCTGGTTCAGTTTCCAGGCCGAGAGGCCCGCGTTGAACGTCGTCATCTTCCCCCTGCTCC .........................................((......)).....((((((((..(((((..(((((((.((..(((((....))..)))..)).))))))).)).)))..)))).))))....................................................................................................................... .........................................42..........................................................................................134.................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................CTCACACCCCGTTAACGGCTGACCTC.......................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CCCCAGGTCTCCCGtctt............................................................................................................................ | 18 | tctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |