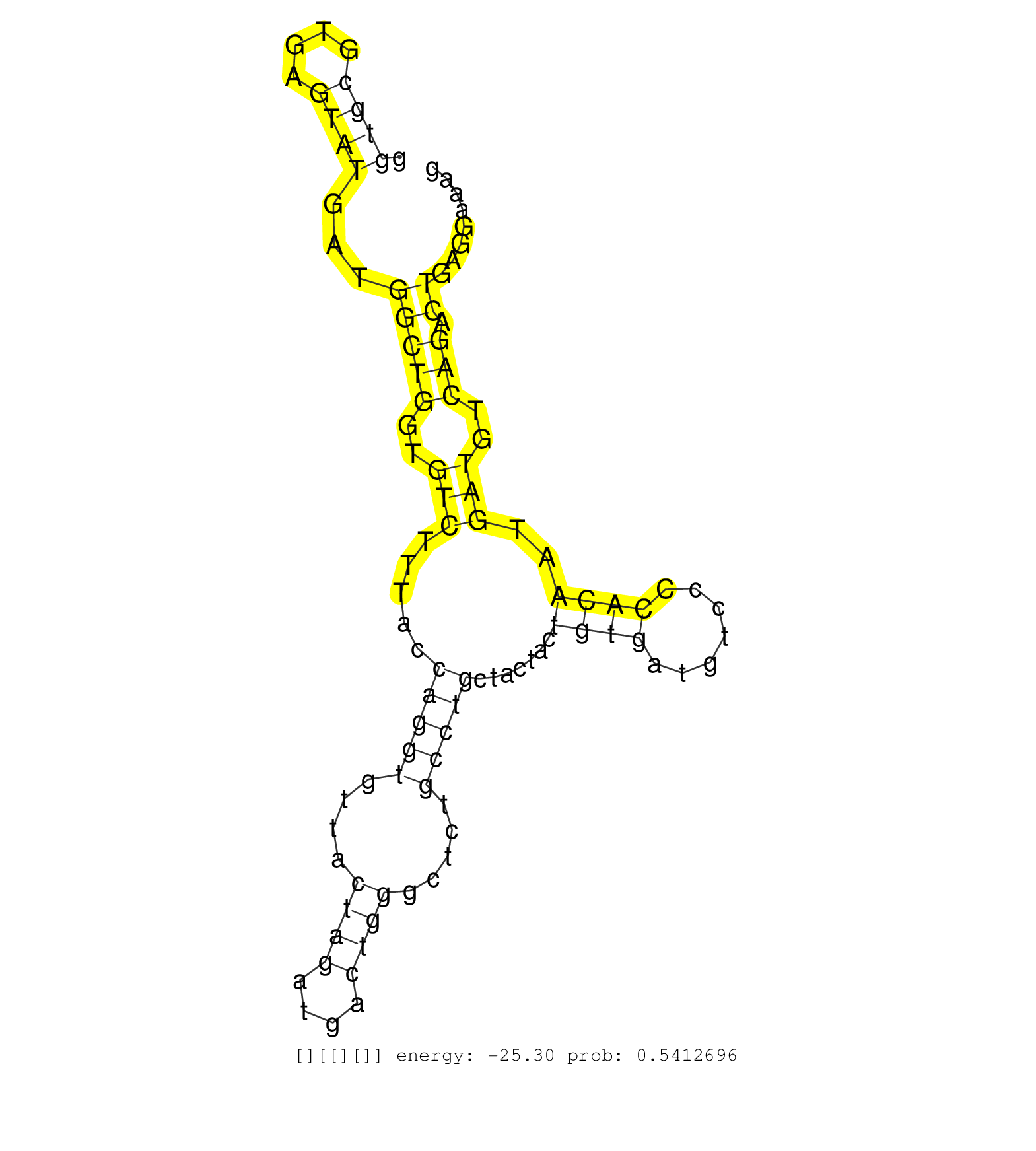

| Gene: D930005D10Rik | ID: uc009ais.1_intron_10_0_chr5_142979905_r.5p | SPECIES: mm9 |
 |
 |
|
(12) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(34) TESTES |
| TCAGGCATCTCCTGCTCAAGATCTCCAAAAGGGTCCGAGACACTGTCTGGGTGCGTGAGTATGATGGCTGGTGTCTTTACCAGGTGTTACTAGATGACTGGGCTCTGCCTGCTACTACTGTGATGTCCCCACAATGATGTCAGACTGAGGAAAGGGCTGGCTTCTCCACATATACCTCTCTACCCAGCAGGCCACCAGGTTGGGGCACTGCAGGCCTCAGTGTAGCATTTAGCCTTCAGAGTGAGTCTGA ..................................................((((....))))...(((((..(((.....(((((....((((....)))).....))))).......((((.......))))..)))..))).))........................................................................................................ .................................................50......................................................................................................154.............................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................CCACAATGATGTCAGACTGAGG.................................................................................................... | 22 | 1 | 426.00 | 426.00 | 113.00 | 116.00 | 86.00 | 39.00 | 15.00 | 7.00 | 7.00 | 1.00 | 11.00 | 3.00 | 1.00 | 4.00 | 1.00 | 1.00 | 4.00 | 1.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | 2.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - |
| .................................................................................................................................CACAATGATGTCAGACTGAGG.................................................................................................... | 21 | 1 | 112.00 | 112.00 | 31.00 | 24.00 | 16.00 | 14.00 | 8.00 | 8.00 | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | 3.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGAGGA................................................................................................... | 23 | 1 | 79.00 | 79.00 | 20.00 | 12.00 | 28.00 | 1.00 | 3.00 | 5.00 | 1.00 | 1.00 | - | 4.00 | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................CACAATGATGTCAGACTGAGGA................................................................................................... | 22 | 1 | 15.00 | 15.00 | 4.00 | 2.00 | 5.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGAGGAA.................................................................................................. | 24 | 1 | 14.00 | 14.00 | - | 1.00 | 1.00 | - | 1.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................CACAATGATGTCAGACTGAGGAAA................................................................................................. | 24 | 1 | 7.00 | 7.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CACAATGATGTCAGACTGAGGAA.................................................................................................. | 23 | 1 | 6.00 | 6.00 | - | 1.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGAG..................................................................................................... | 21 | 1 | 5.00 | 5.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGAGGAAA................................................................................................. | 25 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGCTCAAGATCTCCAAAAGGGTCCGAGA.................................................................................................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGAcg.................................................................................................... | 22 | cg | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CACAATGATGTCAGACTGAG..................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CCCACAATGATGTCAGACTGAGG.................................................................................................... | 23 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGA...................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGcgg.................................................................................................... | 22 | cgg | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGTCAGACTGAGGAAAGGGCTGGC......................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CACAATGATGTCAGACTGAGGc................................................................................................... | 22 | c | 1.00 | 112.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGAGt.................................................................................................... | 22 | t | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CACAATGATGTCAGACTtagg.................................................................................................... | 21 | tagg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGCTCAAGATCTCCAAAAGGGTCCGAGAC................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TCTCCAAAAGGGTCCGAGACACTGTC........................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................CCCACAATGATGTCAGACTGAGt.................................................................................................... | 23 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CACAATGATGTCAGACTGAGGg................................................................................................... | 22 | g | 1.00 | 112.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CCCACAATGATGTCAGACTGAGGA................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTG....................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................ACAATGATGTCAGACTGAGGA................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CACAATGATGTCAGACTG....................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CACAATGATGTCAGACTGAGGAAAGGGaa............................................................................................ | 29 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGAGGAAg................................................................................................. | 25 | g | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCACAATGATGTCAGACTGAGta................................................................................................... | 23 | ta | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................GTGAGTATGATGGCTGGTGTCTTT............................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GTCCGAGACACTGTCTGG........................................................................................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCAGGCATCTCCTGCTCAAGATCTCCAAAAGGGTCCGAGACACTGTCTGGGTGCGTGAGTATGATGGCTGGTGTCTTTACCAGGTGTTACTAGATGACTGGGCTCTGCCTGCTACTACTGTGATGTCCCCACAATGATGTCAGACTGAGGAAAGGGCTGGCTTCTCCACATATACCTCTCTACCCAGCAGGCCACCAGGTTGGGGCACTGCAGGCCTCAGTGTAGCATTTAGCCTTCAGAGTGAGTCTGA ..................................................((((....))))...(((((..(((.....(((((....((((....)))).....))))).......((((.......))))..)))..))).))........................................................................................................ .................................................50......................................................................................................154.............................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................GAGACACTGTCTGGGagt....................................................................................................................................................................................................... | 18 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................GAGACACTGTCTGGGTGaga..................................................................................................................................................................................................... | 20 | aga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |