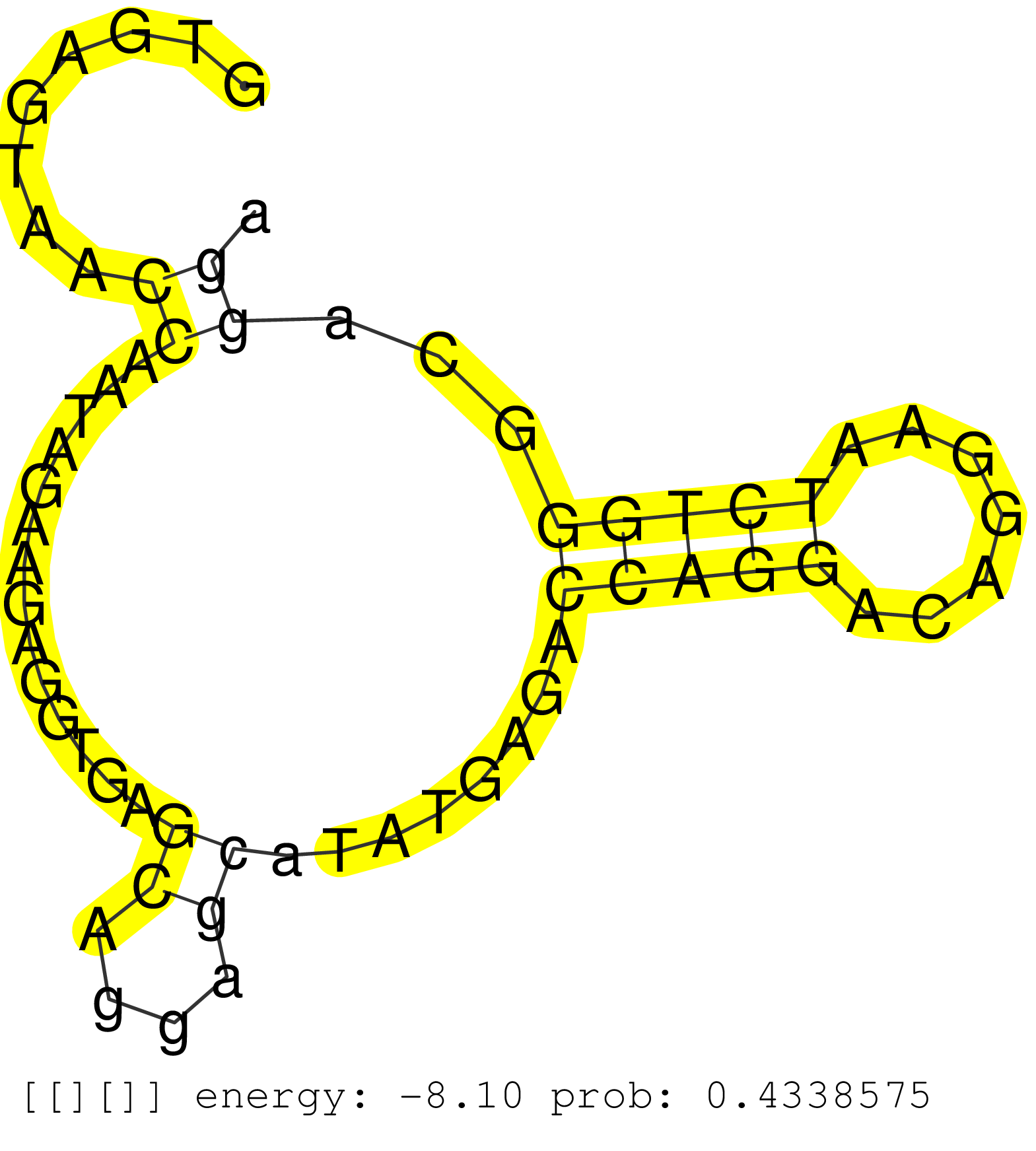

| Gene: Unc84a | ID: uc009agb.1_intron_18_0_chr5_139717982_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| GTACTCCCAAGACAAGACGGGGATGGTGGACTTTGCTCTGGAGTCTGGAGGTGAGTAACCAATAGAAGAGGTGAGCAGGAGCATATGAGACCAGGACAGGAATCTGGGCAGGACTACAAGACACTGCCACCCGTTCCCACTACCCTCAGGGTTTTCTTGTGTAGTCCTGGCTGGCCTGGAACTGAATCTGTAGACCAGGCTATCCTCAATCTCAGATATCCTTCTGTCTCTGCCTCCCAAGTCTGGGATA ..........................................................((..............((....))........(((((.......)))))...)).......................................................................................................................................... ..................................................51............................................................113....................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................TATGAGACCAGGACAGGAATCTGGGC............................................................................................................................................. | 26 | 1 | 8.00 | 8.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGTAACCAATAGAAGAGGTGAGCA............................................................................................................................................................................. | 27 | 1 | 6.00 | 6.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGAACTGAATCTGTAGACCAGGCTA................................................ | 26 | 1 | 6.00 | 6.00 | - | - | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................TCTGGAGTCTGGAGGTGgcag................................................................................................................................................................................................. | 21 | gcag | 4.00 | 0.00 | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGAACTGAATCTGTAGACCAGGCTAT............................................... | 27 | 1 | 4.00 | 4.00 | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................TTTGCTCTGGAGTCTGGAGGTGgca.................................................................................................................................................................................................. | 25 | gca | 4.00 | 0.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGAACTGAATCTGTAGACCAGGCTATCC............................................. | 29 | 1 | 3.00 | 3.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................TGAGACCAGGACAGGAATCTGGGCAGG.......................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGAATCTGTAGACCAGGCTATCCTCAAT........................................ | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................GAATCTGTAGACCAGGCTATCCTCAATC....................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................ATGAGACCAGGACAGGAATCTGGGC............................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTAACCAATAGAAGAGGTGAGC.............................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGAATCTGTAGACCAGGCTATCCTCAA......................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CTTTGCTCTGGAGTCTGGAGGTGgcag................................................................................................................................................................................................. | 27 | gcag | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTAACCAATAGAAGAGGTGAG............................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTAACCAATAGAAGA..................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......CCAAGACAAGACGGGGATGGTGGAC........................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................ACGGGGATGGTGGACTTTGCTCTG.................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CTGAATCTGTAGACCAGGCTATCCTC........................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CCCAAGACAAGACGGGGATGGTGGAC........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................................................................................TGAATCTGTAGACCAGGCTATCCTCAATCT...................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CCCAAGACAAGACGGGGATGGTGGACa.......................................................................................................................................................................................................................... | 27 | a | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TTGTGTAGTCCTGGCTGGCCTGGtt..................................................................... | 25 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TGAGACCAGGACAGGAATCTGGGCAG........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ACTGAATCTGTAGACCAGGCTATCCTCAATC....................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TGAGACCAGGACAGGAATCTGGGCAGGAC........................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................................................TGAATCTGTAGACggg.................................................... | 16 | ggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....TCCCAAGACAAGACGGGGATGGTGGAC........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGAACTGAATCTGTAGACCAGGC.................................................. | 24 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TATGAGACCAGGACAGGAATa.................................................................................................................................................. | 21 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGAATCTGTAGACCAGGCTATCCTCAATC....................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................GAGACCAGGACAGGAATCTGGGCAG........................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCTGGGCAGGACTACAAGACACTGCCACC....................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGAACTGAATCTGTAGACCAGGCT................................................. | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| ............................................................................................................................................................TTGTGTAGTCCTGGCTGGCCTGGAAC.................................................................... | 26 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TTTTCTTGTGTAGTCCTGGCTG............................................................................. | 22 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTACTCCCAAGACAAGACGGGGATGGTGGACTTTGCTCTGGAGTCTGGAGGTGAGTAACCAATAGAAGAGGTGAGCAGGAGCATATGAGACCAGGACAGGAATCTGGGCAGGACTACAAGACACTGCCACCCGTTCCCACTACCCTCAGGGTTTTCTTGTGTAGTCCTGGCTGGCCTGGAACTGAATCTGTAGACCAGGCTATCCTCAATCTCAGATATCCTTCTGTCTCTGCCTCCCAAGTCTGGGATA ..........................................................((..............((....))........(((((.......)))))...)).......................................................................................................................................... ..................................................51............................................................113....................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|