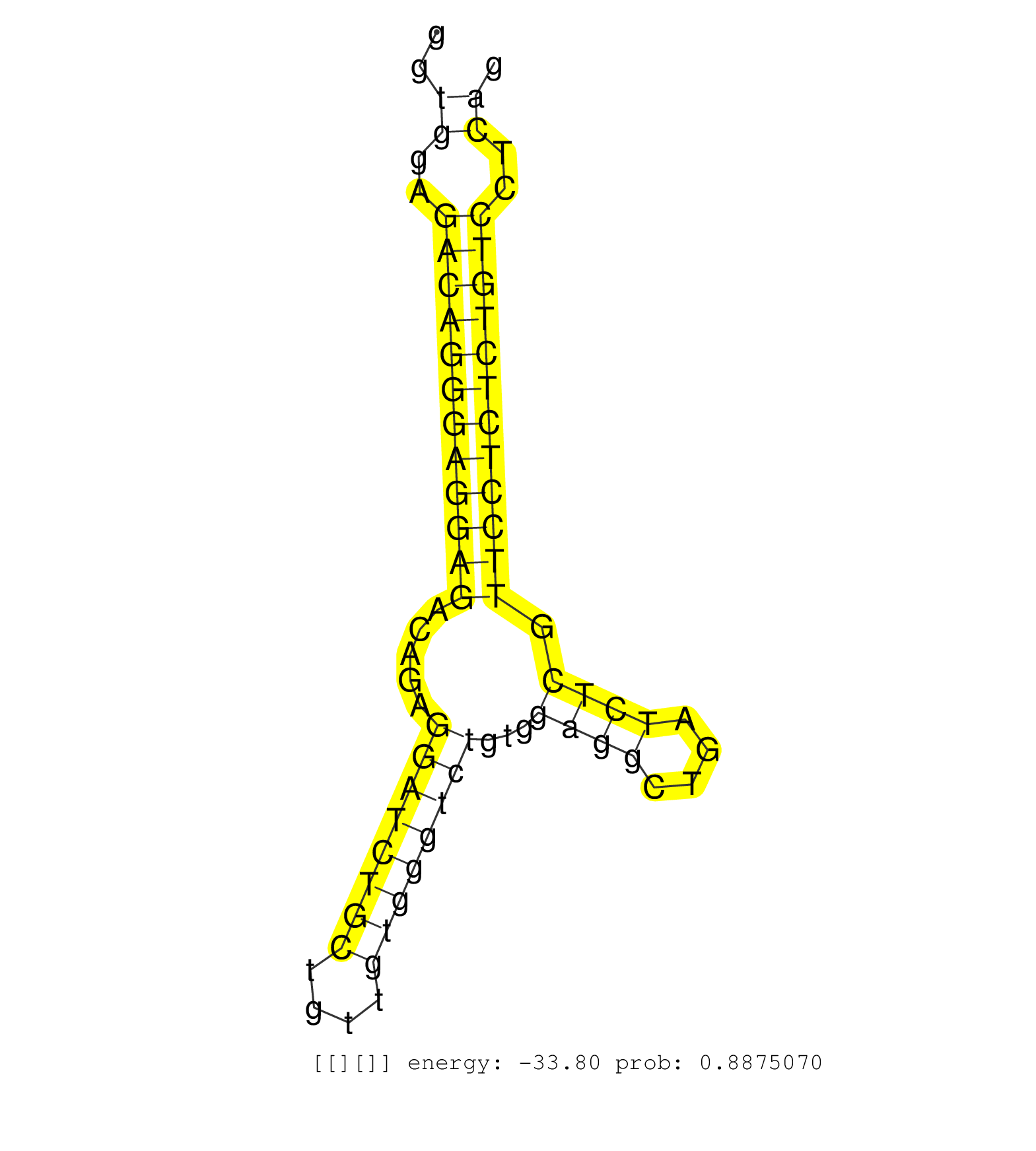
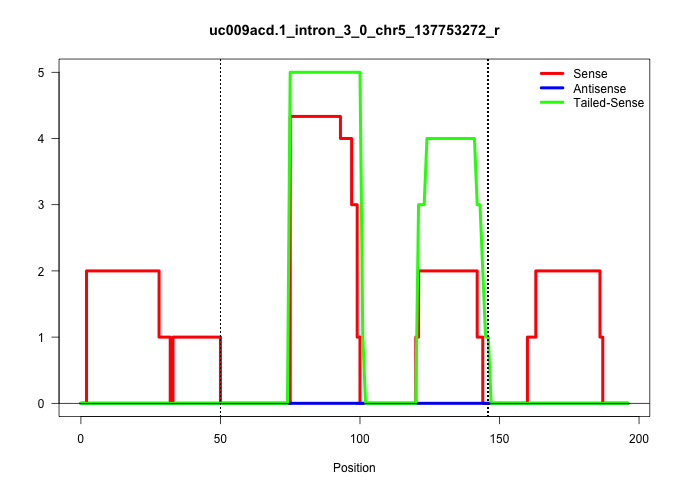
| Gene: Trip6 | ID: uc009acd.1_intron_3_0_chr5_137753272_r | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| CAAGAAACTGGTGCATGACATGAGCCACCCTCCCAGTGGGGAGTACTTTGGTGAGCTGACTGCTGGATAGGGTGGAGACAGGGAGGAGACAGAGGATCTGCTGTTGTGGGTCTGTGGAGGCTGATCTCGTTCCTCTCTGTCCTCAGGTCGGTGTGGTGGCTGTGGCGAAGATGTGGTGGGCGATGGAGCTGGGGTT ........................................................................((..((((((((((((.....((((((((....))))))))...((((....)))).))))))))))))..))................................................... ......................................................................71.........................................................................146................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................TCTCGTTCCTCTCTGTCCTCAG.................................................. | 22 | 1 | 12.00 | 12.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AGACAGGGAGGAGACAGAGGATCTGt............................................................................................... | 26 | t | 4.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGACTGCTGGATAGGGTGGA........................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................GTGTGGTGGCTGTGGCGAA........................... | 19 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TGGTGGCTGTGGCGAAGA......................... | 18 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AGACAGGGAGGAGACAGAGGATCT................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GTGGGCGATGGAGCTGGGG.. | 19 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..AGAAACTGGTGCATGACATGAGCCACCCTC.................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................AGACAGGGAGGAGACAGAGGAT................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................AGACAGGGAGGAGACAGAGGATCTG................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CTGATCTCGTTCCTCTCTGTCCTC.................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGGCTGTGGCGAAGATGTGGTGGGCGATG........... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GTGGCTGTGGCGAAGATGTGGTGGGCGATGG.......... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGATCTCGTTCCTCTCTttcc...................................................... | 21 | ttcc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................CTGTGGCGAAGATGTGGTGGGCGATG........... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................CGGTGTGGTGGCTGTGGCG............................. | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGATCTCGTTCCTCTCTGTCCTat................................................... | 24 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................TGATCTCGTTCCTCTCTGTCCct.................................................... | 23 | ct | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GGCGAAGATGTGGTGGGCGATGG.......... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AGACAGGGAGGAGACAGAGGATCTGCa.............................................................................................. | 27 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGATCTCGTTCCTCTCTGTCC...................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGTGGCGAAGATGTGGTGGGCGATGGA......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................TCTCGTTCCTCTCTGTCCTCAGa................................................. | 23 | a | 1.00 | 12.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..AGAAACTGGTGCATGACATGAGCCAC........................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................CAGTGGGGAGTACTTTG.................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................AGACAGGGAGGAGACAGA....................................................................................................... | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| CAAGAAACTGGTGCATGACATGAGCCACCCTCCCAGTGGGGAGTACTTTGGTGAGCTGACTGCTGGATAGGGTGGAGACAGGGAGGAGACAGAGGATCTGCTGTTGTGGGTCTGTGGAGGCTGATCTCGTTCCTCTCTGTCCTCAGGTCGGTGTGGTGGCTGTGGCGAAGATGTGGTGGGCGATGGAGCTGGGGTT ........................................................................((..((((((((((((.....((((((((....))))))))...((((....)))).))))))))))))..))................................................... ......................................................................71.........................................................................146................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) |
|---|