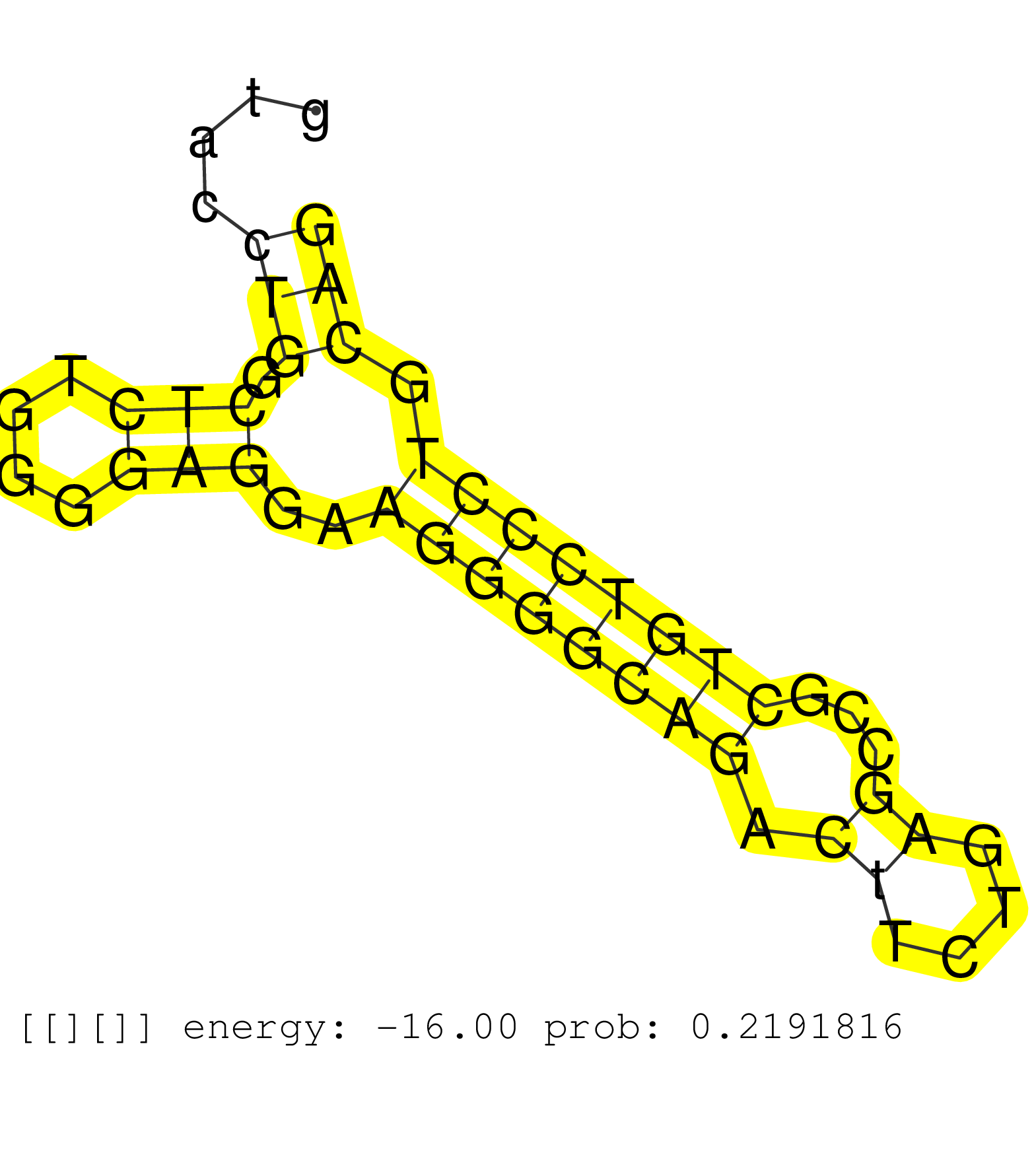
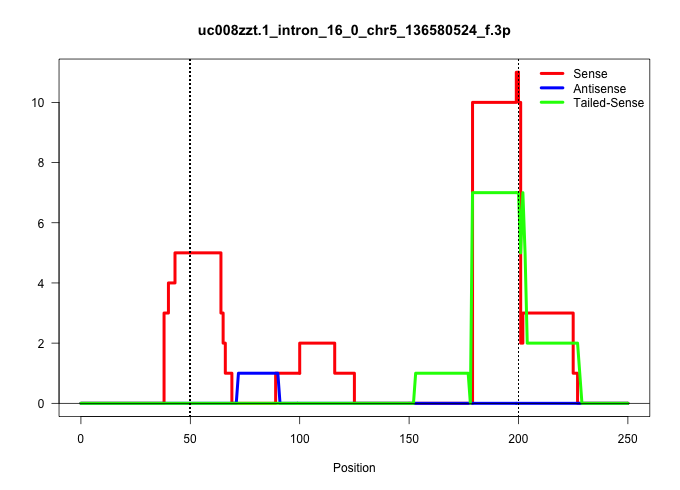
| Gene: Rasa4 | ID: uc008zzt.1_intron_16_0_chr5_136580524_f.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(24) TESTES |
| TCATCAATCAAAAATACCCCACAGGCTTCCCCACAGGTTAATCTGATGGAGACAGCTGCTCATTTGATCACCCCATTCCCAGATTGCTCTAGCTTCTTTGTACCAAGTGGAGAGTCTGTGAGGGGCTGGGAGAGTGGCTCAGAAGAGAGTACCTGGCTCTGGGGAGGAAGGGGCAGACTTCTGAGCCGCTGTCCCTGCAGTGTGTGAACGAGTTGAACCAGTGGCTGTCTGCATTGCGCAAAGCGAGCAC ........................................................................................................................................................(((.(((....)))..((((((((.((....))...)))))))).))).................................................. ....................................................................................................................................................149................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGT................................................. | 22 | 1 | 47.00 | 47.00 | 19.00 | 8.00 | 4.00 | 5.00 | 4.00 | - | 1.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGa................................................. | 22 | a | 21.00 | 18.00 | 14.00 | 3.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAG.................................................. | 21 | 1 | 18.00 | 18.00 | 4.00 | 2.00 | 8.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGTa................................................ | 23 | a | 4.00 | 47.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCA................................................... | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TAATCTGATGGAGACAGCTGCTCATT.......................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTGAACGAGTTGAACCAGTGGCTG....................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGTta............................................... | 24 | ta | 1.00 | 47.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TGATGGAGACAGCTGCTCATTTGATC..................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TGGCTCTGGGGAGGAAGGGGCAGAa........................................................................ | 25 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................TACCAAGTGGAGAGTCTGTGAGGGG............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTGAACGAGTTGAACCAGTGGCTGgg..................... | 27 | gg | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGTagt.............................................. | 25 | agt | 1.00 | 47.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................TAGCTTCTTTGTACCAAGTGGAGAGTC...................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGTata.............................................. | 25 | ata | 1.00 | 47.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGTaaa.............................................. | 25 | aaa | 1.00 | 47.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGTG................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGTat............................................... | 24 | at | 1.00 | 47.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GTGTGTGAACGAGTTGAACCAGTGGC......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TGTGTGAACGAGTTGAACCAGTGGC......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTGAACGAGTTGAACCAGTGGCTGg...................... | 26 | g | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TAATCTGATGGAGACAGCTGCTCATTT......................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGAGCCGCTGTCCCTGCAGTt................................................ | 23 | t | 1.00 | 47.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATCTGATGGAGACAGCTGCTCATTTG........................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCATCAATCAAAAATACCCCACAGGCTTCCCCACAGGTTAATCTGATGGAGACAGCTGCTCATTTGATCACCCCATTCCCAGATTGCTCTAGCTTCTTTGTACCAAGTGGAGAGTCTGTGAGGGGCTGGGAGAGTGGCTCAGAAGAGAGTACCTGGCTCTGGGGAGGAAGGGGCAGACTTCTGAGCCGCTGTCCCTGCAGTGTGTGAACGAGTTGAACCAGTGGCTGTCTGCATTGCGCAAAGCGAGCAC ........................................................................................................................................................(((.(((....)))..((((((((.((....))...)))))))).))).................................................. ....................................................................................................................................................149................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................CCATTCCCAGATTGCTCTA............................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................CAGTGGCTGTCTGCAagg................. | 18 | agg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |