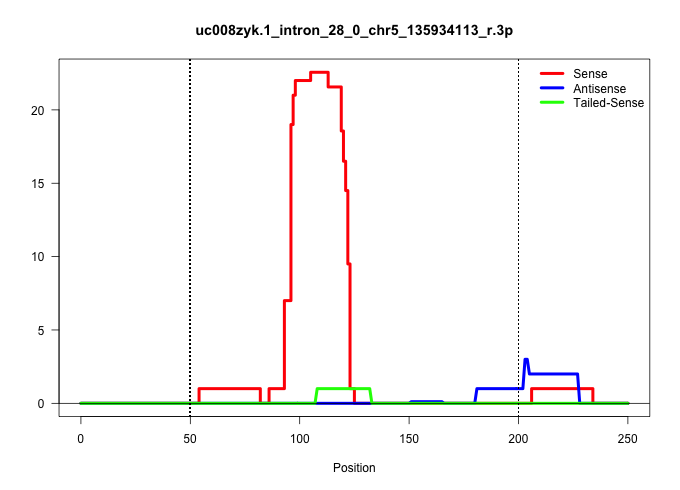
| Gene: Hip1 | ID: uc008zyk.1_intron_28_0_chr5_135934113_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) PIWI.mut |
(11) TESTES |
| TAATTTAAAATGTAATTAAAAAGAAAGAATTCTTTTTCAGGGGAAACTGTTATAAAATAGGTAACGAGGTATTTATACCCTCAGCCTCCAAGCTGCTAGGCAAACGTGTAGAACTGTGGGGTCTTAGCCATGAGCTGAGCTCCGCCTCCCCCTTACCCAGCTCCACCCACCAAGCCCTTGTCTTCCTTGACTCCTCCCACCGTGTATTCTGGGCACCCACCATGAGAAAGGAGCGCAGACCTTTTGGTCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................TAGGCAAACGTGTAGAACTGTGGGGTC............................................................................................................................... | 27 | 1 | 7.00 | 7.00 | 2.00 | 1.00 | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 |
| ................................................................................................TAGGCAAACGTGTAGAACTGTGGGGT................................................................................................................................ | 26 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - |
| .............................................................................................TGCTAGGCAAACGTGTAGAACTGTGG................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGCTAGGCAAACGTGTAGAACTGTGGG.................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TTCTGGGCACCCACCATGAGAAAGGAGC................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGCTAGGCAAACGTGTAGAACTGTGGGG................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AGGCAAACGTGTAGAACTGTGGGGT................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................TCCAAGCTGCTAGGCAAACGTGTAGAA......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................AGGCAAACGTGTAGAACTGTGGGGTC............................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................GGCAAACGTGTAGAACTGTGGGGTCTT............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TAGAACTGTGGGGTCTTAGCCATGt..................................................................................................................... | 25 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................AAATAGGTAACGAGGTATTTATACCCTC........................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................TAGGCAAACGTGTAGAACTGTGGGG................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................GTGTAGAACTGTGGGGTC............................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - |
| .........................................................................................................GTGTAGAACTGTGGG.................................................................................................................................. | 15 | 16 | 0.06 | 0.06 | - | - | 0.06 | - | - | - | - | - | - | - | - |
| TAATTTAAAATGTAATTAAAAAGAAAGAATTCTTTTTCAGGGGAAACTGTTATAAAATAGGTAACGAGGTATTTATACCCTCAGCCTCCAAGCTGCTAGGCAAACGTGTAGAACTGTGGGGTCTTAGCCATGAGCTGAGCTCCGCCTCCCCCTTACCCAGCTCCACCCACCAAGCCCTTGTCTTCCTTGACTCCTCCCACCGTGTATTCTGGGCACCCACCATGAGAAAGGAGCGCAGACCTTTTGGTCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................GTATTCTGGGCACCCACCATGAGAA...................... | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CTTCCTTGACTCCTCCCACCGTGT............................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................CTTACCCAGCTCCAC.................................................................................... | 15 | 9 | 0.11 | 0.11 | - | - | 0.11 | - | - | - | - | - | - | - | - |